Based on Mendelian genetics, it is possible to predict the probability of the appearance of a particular allele in an offspring when the alleles of each parent are known. Similar predictions can be made about the frequencies of alleles in the next generation of an entire population. By comparing the predicted or "expected" frequencies with the actual or "observed" frequencies in a real population, scientists can infer a number of possible external factors that may be influencing the genetic structure of the population (such as inbreeding or selection).
Hardy-Weinberg Principle
Based on Mendel's principles of inheritance, two men, G.H. Hardy and Wilhelm Weinberg, independently developed the concept that is known today as the Hardy-Weinberg Principle, which states: "In a large, randomly breeding (diploid) population, allelic frequencies will remain the same from generation to generation; assuming no unbalanced mutation, gene migration, selection or genetic drift." When a population meets all of the Hardy- Weinberg conditions it is said to be in Hardy-Weinberg equilibrium . This equilibrium can be mathematically expressed based on simple binomial (for two alleles) or multinomial (multiple allele) distribution of the gene frequencies as above: 
Testing for Hardy-Weinberg Equilibrium
Populations in their natural environment can never meet all of the conditions required to achieve Hardy-Weinberg equilibrium, thus their allele frequencies will change from one generation to the next and the population will evolve. Just how far the population deviates from Hardy-Weinberg is an indication of the intensity of external factors, and can be determined by a statistical formula called a  (chi-squared), which is used to compare observed versus expected outcomes:
(chi-squared), which is used to compare observed versus expected outcomes:

Where:
- O is the observed number of individuals with the specific genotype, and E is the expected number of individuals based on the Hardy-Weinberg Equilibrium (i.e., for AA, the expected number of individuals is:
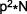 )
)
- k is the number of genotypes
Example continued from Hardy-Weinberg discussion above:

 is used to determine the probability that the observed number differs from the expected number due to chance alone. Standardized statistical charts have been developed which correlate the
is used to determine the probability that the observed number differs from the expected number due to chance alone. Standardized statistical charts have been developed which correlate the  value and degrees of freedom (the number of independent variables) with probability values (p). In the above example, the
value and degrees of freedom (the number of independent variables) with probability values (p). In the above example, the  value of 27.77 correlates to a greater than 99% chance that the difference between the observed and the expected is NOT due to chance alone. This high probability indicates that some external factor (i.e., migration, selection, inbreeding, or drift) is influencing the frequencies of alleles.
value of 27.77 correlates to a greater than 99% chance that the difference between the observed and the expected is NOT due to chance alone. This high probability indicates that some external factor (i.e., migration, selection, inbreeding, or drift) is influencing the frequencies of alleles.
 is a statistical calculation used not just for population genetics, but for comparing any pair of data to determine if there is an association between them.
is a statistical calculation used not just for population genetics, but for comparing any pair of data to determine if there is an association between them.
Effective Population Size
One of the many variables of population dynamics that can influence the rate and size of fluctuation in allele frequencies is population size. Genetic drift , the random increase or decrease of an allele's frequency, affects small populations more severely than large ones, since alleles are drawn from a smaller parental gene pool. The rate of change in allele frequencies in a population is determined by the population's effective population size. The effective population size is the number of individuals that evenly contribute to the gene pool.
The actual number of individuals in a population is rarely the effective population size. This is because some individuals reproduce at a higher rate than others (have a higher fitness), the distribution of males and females may result in some individuals being unable to secure a mate, or inbreeding reduces the unique contribution of an individual. The effective population size is a theoretical measure that compares a population's genetic behavior to the behavior of an "ideal" population. As the effective population size becomes smaller, the chance that allele frequencies will shift due to chance (drift) alone becomes greater.
There are many formulas for calculating effective population size (  ). One that takes into consideration the number of breeding males and females, and their sex ratio, is:
). One that takes into consideration the number of breeding males and females, and their sex ratio, is:

Application of this formula requires the same assumptions as the Hardy-Weinberg equilibrium. There are other, more complicated versions of the formula which can take into account exceptions such as inbreeding and fitness.
Inbreeding and Relatedness
Small effective population size can result in a high occurrence of inbreeding, or mating between close relatives. One of the effects of inbreeding is a decrease in the heterozygosity (increase in homozygosity) of the population as a whole, which means a decrease in the number of heterozygous genes in the individuals. This effect places individuals and the population at a greater risk from homozygous recessive diseases that result from inheriting a copy of the same recessive allele from both parents. The impact of accumulating deleterious homozygous traits is called inbreeding depression - the loss in population vigor due to loss in genetic variability or genetic options.
In the 1950's, Sewell Wright developed a set of parameters called F statistics. The simplest of these is the inbreeding coefficient defined as the probability that two homologous (same) alleles present in the same individual are identical by descent. The inbreeding coefficient (F) is calculated by comparing the expected heterozygosity 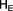 with observed heterozygosity (
with observed heterozygosity (  ), and ranges from -1 (no inbreeding) to +1 (complete identity).
), and ranges from -1 (no inbreeding) to +1 (complete identity).

If the values for both observed and expected heterozygosity are the same, F will be zero. A positive value indicates that there is an increased number of homozygotes, and population may be inbred - the larger the number, the greater the extent of inbreeding. A negative value indicates that there are more heterozygous individuals than would be expected; this might happen for the first few generations after two previously isolated populations become one.
In the  example above:
example above: 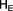 = 10 and
= 10 and  = 24.368 for the heterozygote, thus
= 24.368 for the heterozygote, thus
F = (10 - 24.368) / 10 = -1.43
We determined earlier (using  ) that the difference between the observed and expected is not likely due to chance. However, since there is an increase in the expected number of heterozygotes, inbreeding can be ruled out as a possible population dynamic that is influencing the genotype frequencies.
) that the difference between the observed and expected is not likely due to chance. However, since there is an increase in the expected number of heterozygotes, inbreeding can be ruled out as a possible population dynamic that is influencing the genotype frequencies.
Wright's Coefficient of Relationship (RC)
The degree to which inbreeding has occurred can be mathematically calculated from pedigree analysis. Wright's Coefficient of Relationship (RC) is the probability that homologous alleles present in different individuals are identical by descent.
The degree to which inbreeding has occurred can be mathematically calculated from pedigree analysis. Wright's Coefficient of Relationship (RC) is the probability that homologous alleles present in different individuals are identical by descent.
In a diploid organism, offspring inherit ½ of their alleles from a single parent. Thus, by following the path of relationships in a pedigree, the RC can be determined from the multiplication of each order of relatedness. For example:

These example relationships are very straight-forward, however relationships between individuals, particularly in domestic and endangered species, can be much more complex including crosses between siblings, and backcrosses between grand-offspring and grandparents. For complex pedigrees, the overall RC can calculated based on a collective of the individual RC's by:

Where:
 is the sum
is the sum  of all paths connecting X and Y, and indicates the probability that any gene is homozygous by descent.
of all paths connecting X and Y, and indicates the probability that any gene is homozygous by descent. - N is the number of connections between X and Y through each common ancestor.
 is the inbreeding co-efficient of the ancestor.
is the inbreeding co-efficient of the ancestor.  and
and  are the inbreeding coefficients for X and Y.
are the inbreeding coefficients for X and Y.
Out-breeding Depression
In some cases, inbreeding is purposely done to create a "pure line" of individuals which all have similar genes. This is the case with many domesticated species such as pure-bred pets, laboratory animals, and agricultural plants and animals. Breeders intentionally inbreed to create a population that has the genes for desired traits in a homozygous state, so that the offspring will perpetuate the desired phenotype. Over generations, individuals with deleterious genes are removed from the breeding population, resulting in a monomorphic, yet healthy population. These populations can actually suffer "out-breeding depression" when crossed to unrelated individuals which results in new combinations of alleles, and possible entry of deleterious genes.
Inbreeding or "line-breeding" as it is referred to by pet breeders, must be done very carefully to prevent the creation of a line that is almost perfect but contains a major flaw, such as domestic dog breeds that have the propensity for hearing problems (Dalmatians, cocker spaniels) or seizures (Irish setters). Responsible breeders will often discontinue breeding a line whenever the slightest chance of a deleterious gene is noticed, sacrificing many years of work to prevent damaging the breed.