|
|
|
 |
|
|
|
|
A biosystem, or biological system, is a group of molecules that interact in a biological system. One type of biosystem is a biological pathway, which can consist of interacting genes, proteins, and small molecules. Another type of biosystem is a disease, which can involve components such as genes, biomarkers, and drugs.
An understanding of the components, products, and biological effects of biosystems can lead to better understanding of biological processes in normal and disease states, elucidation of possible drug effects and side effects, and other insights to complex processes that have implications for health and medicine.
The COX portion of the Arachidonic acid metabolism pathway, for example, metabolizes lipids commonly found in the cell membrane into prostaglandins, which are involved in regulation of neuronal activity. This pathway has an important role in pain and inflammation. Specifically, the protein encoded by human PTGS1 gene is involved in the conversion of prostaglandin PGG2 into inflammation-causing prostaglandin PGH2. Aspirin has been shown to bind to the PTGS1 gene product (prostaglandin-endoperoxide synthase 1), blocking that enzyme's ability to produce PGH2 and thereby reducing pain and inflammation.
To illustrate the ways in which the NCBI BioSystems database can be used, the Arachidonic acid metabolism pathway is used in illustrations and examples threaded through this help document.
|
|
|
|
|
|
 |
|
|
|
|
 
Numerous publicly browsable biosystems databases are accessible throughout the Web. The NCBI BioSystems database was designed to centralize access to those existing databases and to integrate their records with associated data types (genes, proteins, small molecules, etc.) as noted below. The current source databases are listed in the section on "content of the BioSystems database."
 
Upon submission to the NCBI BioSystems database, records from source databases go through an automated data processing pipeline to identify associated literature, molecular, and chemical data throughout the Entrez system, and to then establish connections among those data sets. This allows seamless traversal from a biosystem record to (a) sequence, 3D structure, and chemical data for the biosystem components; (b) PubMed records for articles about the biosystem; and (c) additional information associated with the biosystem components, such as functional and evolutionary classification of protein components in the Conserved Domain Database (CDD), 3D protein structures in the Molecular Modeling Database (MMDB), biological activity data for component small molecules in the PubChem BioAssay database, and phenotypic information in OMIM.
 
Many biosystem databases provide detailed diagrams that permit close examination of the components and biochemical steps within individual pathways. The NCBI BioSystems database was developed as a complementary project to list and categorizes the genes, proteins, and small molecules involved in each biological system, and to facilitate computation on biosystems data.
For example, using the human Arachidonic acid metabolism pathway (mentioned in "what is a biosystem?") as an illustration, the NCBI BioSystems database can be used to answer such questions as the following:
What genes, proteins, and small molecules are involved in this pathway? Where in the metabolic pathway do my gene(s), protein(s), or small molecule(s) of interest occur? What biosystems include a particular component of interest?
Example 1: To retrieve biosystems that include the human PTGS1 gene as a component, you can search the GeneID field of the BioSystems database for that component's unique identifier (in this case, 5742). The biosystems that are retrieved have been associated with that gene through the method descibed in the data processing section of this document. Example 2: To retrieve biosystems that include prostaglandin G2 (PGG2, CID 5280883) as a component, you can search the Compound ID (CID) field of the BioSystems database for that component's unique identifier (in this case, 5280883). The biosystems that are retrieved have been associated with that CID through the method descibed in the data processing section of this document.
In what other organisms is this pathway found?
|
|
|
|
|
|
 |
|
|
|
|
 
The content of the BioSystems database currently comes from the following sources. Articles cited in the References section provide more information about each of the source databases.
|
KEGG: Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/) by the Kanehisa Laboratory of the Bioinformatics Center, Institute for Chemical Research, Kyoto University, Kyoto, Japan. (read more; retrieve KEGG biosystem records) |
|
|
BioCyc (http://biocyc.org/) is a collection of organism-specific pathway/genome databases (PGDBs). The Tier 1 (EcoCyc (http://ecocyc.org/) and MetaCyc (http://metacyc.org/)) and Tier 2 data sets are included in the NCBI BioSystems database. (read more; retrieve BioCyc biosystem records) |
|
|
Pathway Interaction Database (PID, http://pid.nci.nih.gov/) "is a freely available collection of curated and peer-reviewed pathways composed of human molecular signaling and regulatory events and key cellular processes." (read more; retrieve PID biosystem records) |
|
|
Reactome (http://www.reactome.org/) is a curated knowledge base of human biological pathways and inferred equivalent reactions in multiple non-human species. (read more; retrieve Reactome biosystem records) |
|
|
WikiPathways (http://www.wikipathways.org/index.php/WikiPathways) is an open, public platform dedicated to the curation of biological pathways by and for the scientific community. (read more; retrieve WikiPathways biosystem records) |
|
|
The Gene Ontology (GO) project (http://www.geneontology.org/) is an initiative to standardize the representation of gene and gene product attributes across species and databases and provides a controlled vocabulary of terms for describing gene product characteristics and gene product annotation data. (read more)
Retrieve all GO records from the NCBI BioSystems database, or only the:
biological processes (root record)
 These are categorized in the BioSystems database as "pathway"[BioSystemType].
cellular components (root record)
These are categorized in the BioSystems database as "pathway"[BioSystemType].
cellular components (root record)
 These are categorized in the BioSystems database as "structural complex"[BioSystemType].
molecular functions (root record)
These are categorized in the BioSystems database as "structural complex"[BioSystemType].
molecular functions (root record)
 These are categorized in the BioSystems database as "functional set"[BioSystemType].
These are categorized in the BioSystems database as "functional set"[BioSystemType].
If you open the root record for any category, you can use the "Related BioSystems:Subset BioSystems" folder tab to view the nodes beneath it. The other GO records will also have "Subset and/or Superset BioSystems" folder tabs, allowing you to browse up and down the GO hierarchy and to retrieve the associated genes and proteins, as available, for any node.
As noted in "data processing: create direct links" section of this help document, the BioSystems database uses the GO-Lite data files, plus the list of gene<->GO associations provided in the "gene2go" data file of the Entrez Gene FTP site, when creating links from GO terms to genes and proteins.
|
|
|
LIPID Metabolites And Pathways Strategy (LIPID MAPS, http://www.lipidmaps.org/) is a multi-institutional effort to identify and quantitate, using a systems biology approach and sophisticated mass spectrometers, the lipid species in mammalian cells, and to quantitate the changes in these species in response to perturbation. (read more; additional articles; retrieve LIPID MAPS biosystem records) |
|
The content and of BioSystems database records reflect the data provided by the source, and some special functions may vary by source as well. For example, thumbnail images of biosystem diagrams and the ability to highlight selected components in a full size image of the biosystem diagram are currently available for KEGG records. The central functions of the BioSystems database, however, are available for all records, such as the listing and categorization of biosystem components and links to related data throughout the Entrez system.
If you want to limit search results to records from a particular source, you can use the SourceName field, as described in the Search Fields section of this help document.
If you are interested in depositing data into the BioSystems database, please contact biosystems.help@ncbi.nlm.nih.gov.
 
Several types of records are currently available in the BioSystems database:
|
A series of related biochemical reactions. The "pathway" category of records in the NCBI BioSystems database also includes biological processes from Gene Ontology (GO).
Some source databases, such as Gene Ontology (GO), KEGG, and Reactome, organize pathways hierarchically into supersets and subsets (KEGG refers to its subsets as "pathway modules"). The BioSystems database includes both supersets and subsets and provides cross-references between them.
Retrieve all pathway records from the NCBI BioSystems database.
 |
|
|
Biomolecules and/or chemicals that are bound to each other as asserted by the source database. The "structural complex" category of records in the NCBI BioSystems database also includes cellular components from Gene Ontology (GO).
Retrieve all structural complex records from the NCBI BioSystems database.
|
|
|
A group of enzymes that perform a certain function, for example, all aminoacyl transferases. The "functional set" category of records in the NCBI BioSystems database also includes molecular functions from Gene Ontology (GO).
Retrieve all functional set records from the NCBI BioSystems database.
|
|
|
A group of genes that have a pathogenicity or other phenotype associated with them.
Retrieve all signature module records from the NCBI BioSystems database.
|
|
Note: If you want to restrict your search results to a particular type of record, you can select the desired record type using the Limits option on the BioSystems search page, or you can use the "BioSystemType" Search Field. For example, you can add the following term and search field to your query:
yourtermshere AND "pathway"[BioSystemType] or
yourtermshere AND "pathway"[Type]
You can enter any of the record types above as an alternative to "pathway," and you can spell the full name of the search field [BioSystem Type] or just enter [Type] for short.
BioSystem records can either be specific to a single organism, or represent canonical biosystems across a wider taxonomic span:
- organism specific biosystem
|
an organism-specific biosystem that typically includes genes, proteins, and small molecules as components. (If similar biosystems are found in other organisms, you can retrieve them by opening the corresponding conserved biosystem record, accessible under the Related BioSystems folder tab, and then viewing the set of organism-specific biosystems that fall under the conserved biosystem.) |
|
|
canonical biosystems that are used to group together orthologous, organism specific biosystems. These records are derived from "reference pathways" in the KEGG database and from MetaCyc records. The organism specific biosystems that fall under a canonical biosystem are accessible under that record's Related BioSystems folder tab. (Note: The folder tabs in conserved biosystem records may include genes and proteins from multiple organisms in which the biosystem is found. For example, a biosystem record derived from a KEGG reference pathway shows a concatenated list of the genes and proteins present in the organism-specific biosystems associated with the conserved biosystem. To see only the genes and proteins of a single organism�s biosystems, click on the organism of interest in the Related BioSystems/Organism Specific BioSystems folder tab.) |
|
For example, the conserved biosystem for arachidonic acid metabolism, bsid366, encompasses numerous organism-specific biosystems, including bsid82991 for human arachidonic acid metabolism.
If you want to limit your search results to a records with a particular taxonomic scope, you can either select the desired set using the Limits folder tab on the BioSystems search interface, or use the Filter field as described in the Search Fields section of this help document (e.g., add the following term and search field to your query: AND "conserved biosystem"[Filter]).
 
Extract descriptive text: When biosystems records are deposited by a source database, an archival copy of each record is made and text blocks such as biosystem name, description, and comments are then extracted into the BioSystems database.
Extract thumbnail images: If a source database provides a thumbnail image of a biosystem diagram, the image is placed in the corresponding BioSystems database record and links to a full-size image that resides on the source database web site.
- At present, thumbnail images and the ability to highlight selected components (genes, proteins, small molecules) in a full size image of the biosystem diagram are currently available only for KEGG records. In BioSystems records that do not contain a thumbnail image, a full size diagram may be available at the source database by following the link in the BioSystem record for the source's accession number.
|
|
Create direct links: Each biosystem record has one-to-one relationships with specific records in other Entrez databases, particularly for biosystem components in the Entrez Gene, Protein, and PubChem Compound databases, as well as PubMed articles cited in the biosystem record. Reciprocal links between the biosystem record and these component and literature records are created by parsing the source database's record for the following unique identifiers:
| |
Genes |
GeneIDs present in the source record are parsed out and links are then established to the corresponding records in the Entrez Gene database. The links are made available in both the folder tabs and in the "Related information" menu that appear on individual BioSystem records. If only gene names but not GeneIDs are provided in the source record, those gene names are parsed out and listed as components in the genes folder tab of individual BioSystem records but they are not linked to Entrez Gene records and are therefore not retrieved by the "Related information" : Sequences : Genes menu item. For Gene Ontology (GO) records, the BioSystems database uses the list of genes provided in the GO-Lite data files, plus the list of gene<->GO associations provided in the "gene2go" data file of the Entrez Gene FTP site. (Additional details about GO records in the BioSystems database are provided in the source databases section of this document.) For biosystems from all source databases, links are also established to Gene IDs that correspond to the protein sequence GI numbers mentioned below; for example, if one of those protein GIs is cited directly in a Gene record, a link to that Gene record is made. |
|
| |
| |
Proteins |
Protein GI numbers present in the source record are parsed out and links are then established directly to the corresponding sequence records in the Entrez Protein database. If the source record contains protein accession numbers, the current GI number for each accession is determined and a link to the corresponding protein sequence record is made using the GI number.
In addition, the set of links to protein sequences is expanded in the following ways:
(a) If any GI numbers are for RefSeq records, links to corresponding UniProt/Swiss-Prot records are also made.
(b) if any other record(s) in the Entrez Protein database contains an identical sequence to the one present in the cited GI and also share the same Taxonomy ID (TaxID), links to those identical sequence records are established as well.
Technical note: In the data processing pipeline at NCBI, protein sequence records that contain an identical sequence, regardless of TaxID, are placed in a protein identity group (PIG), and each group is given a stable identification number (PIG ID). For step (b) above, links are made to all PIG members that have the same TaxID as the protein GI cited in the source biosystem record. (The proteins are then listed in the protein folder tab of a biosystem record and sorted by PIG ID so identical sequences can be easily spotted.) |
|
| |
| |
Small Molecules |
Records from source databases are parsed for small molecule identification numbers, including PubChem Compound IDs (CIDs), PubChem Substance IDs (SIDs), and external registry names such as local identifiers assigned to a substance by a the source database. The types of links that are made depend upon the type of identifiers that were found:
If SIDs are present in the source record, links are established to the corresponding PubChem Substance records and to associated CIDs in PubChem Compound.
If CIDs are present in the source record, links to the corresponding PubChem Compound records are made (however, the links are not extended to associated PubChem Substances).
If external registry names are present, those identifiers are mapped to the corresponding SIDs and links are made to those records in PubChem Substance as well as to associated CIDs in PubChem Compound.
|
|
| |
Literature |
If the source record includes PubMed identifiers (PMIDs) for journal articles about the biosystem, the PMIDs are parsed and links are established to the corresponding records in the PubMed database. If the source record contains a text citation but no PMID, the citation is displayed in the folder tabs section of the biosystem record, but no link to PubMed is provided. |
|
| |
| |
Taxonomy |
Depositors provide the Taxonomy ID (TaxID) of the source organism for organism specific biosystems. These TaxIDs are parsed and links to the corresponding information in the NCBI Taxonomy database are then established. Taxonomic information is not extracted from conserved biosystems; however, the individual organisms that are encompassed by a conserved biosystem are listed in the Organism Specific BioSystems folder tab of the conserved biosystem record. |
|
Calculate indirect links: Records that are directly linked to a biosystem may in turn have links to other Entrez databases and records. For example, a biosystem record that includes a small molecule component will have a direct link to the PubChem Compound record for that small molecule. If the PubChem Compound record in turn has a link to information about the molecule's biological activity in the PubChem BioAssay Database, a link between the BioSystem record and the BioAssay record will also be established. These types of connections represent indirect (second generation) links.
Identify relationships among biosystems: Two or more biosystem records are considered related if any one of the following conditions have been met:
- the source database has explicitly stated the biosystems are connected in some way (Linked BioSystem)
- the biosystems share at least one identical protein sequence from the same source organism (Similar BioSystems). In other words, the shared protein is in the same protein identity group (PIG) and has the same Taxonomy ID (TaxID).
- the biosystems have an evolutionary relationship, such as an Organism Specific BioSystem that is an instance of a canonical, Conserved BioSystem. For example, human arachidonic acid metabolism (bsid82991) is an organism-specific instance of the conserved biosystem for arachidonic acid metabolism (bsid366). The conserved biosystem record, in turn, links back to the human and other organism-specific instances of the biosystem, reflecting the relationships specified by the source database, KEGG.
- biosystems that have a functional relationship with each other, either as a Superset of smaller biological processes or a Subset of (or small reaction within) a larger biological process. For example, "Phase 1 - Functionalization of compounds" (bsid105699) is a superset biosystem that includes "COX reactions" (bsid105711) as one of its subset biosystems, reflecting the hierarchical organization of those biosystems specified by the source database, Reactome. The superset/subset relationships reflect the node immediately above/below the biosystem being viewed.
In each case, reciprocal links are created among the related biosystems and are accessible from: (1) the folder tabs on individual biosystem records, and (2) the "Related information" menu that appears in the right hand margin of a biosystem record display.
|
|
|
|
|
|
|
|
 |
|
|
|
|
 
text terms (key words): A wide variety of text terms, such as names of biosystems, genes, proteins, and small molecules can be used to search the BioSystems database. You can also search for other words that might be present in the title, comments, or other text containing fields of a record. Because terminology can vary across records, it can be helpful to include synonyms in your query (e.g., PTGS1 OR "prostaglandin endoperoxide synthase 1"). It is also possible to search for a word stem by using an asterisk (*) as a wild card; for example, arachidon* will retrieve records with terms such as arachidonate, arachidonic, arachidonoyl. The Entrez Help document provides additional information about truncating search terms in this way.
unique identifiers: BioSystem records can be retrieved by searching for the unique identifiers of their components, such as GeneIDs, protein sequence GI numbers or accession numbers, and CIDs, SIDs, or external registry names of small molecules. The retrieved biosystems have been associated with those components through the methods descibed in the data processing/create direct links section of this document.
organism: To retrieve biosystem records for a specific organism or organism group, you can enter its common name (e.g., human) or scientific name (e.g., Homo sapiens), or other taxonomic node (e.g, Primates) in the Organism [orgn] search field. Note that a search of the organism field will retrieve only organism specific biosystems that fall under the specified taxonomic node. To find corresponding conserved biosystems that are present in numerous organisms, simply follow the "related biosystems/conserved biosystems" link from the biosystem record(s) of interest.
database subset: It is possible to retrieve records from a specified database subset. For example, the SourceName search field allows you to retrieve records from a specific source database. The Filter field allows you to limit a search to specific record types or to records that have links to another Entrez database of interest; for example, a search for biosystems_structure[filter] will retrieve biosystem records that have links to the Entrez Structure database, and a search for biosystems_omim[filter] will retrieve biosystem records that have links to the Online Mendelian Inheritance in Man (OMIM) database.
and more... The biosystems database can also be searched by terms that appear in any of the other search fields.
 
A variety of techniques can be used to search the database, offering varying degrees of control over your query. In some cases, they offer alternative ways of executing the same search (as is true for sample searches #4, #5, and #6 below), with each method offering different benefits. The search methods include:
| Method |
Description |
Example |
| Basic Search |
Just enter search terms without specifying search fields, other limits, or Boolean operators.
The "Search Details" box in the right margin of the search results page shows exactly how Entrez parsed and handled your query. If desired, you can edit the query in that box and press the "Search" button to run the modified query.
The "See more..." link a the bottom of the "Search Details" box opens a more detailed display:
- The Query Translation box shows the search strategy used to run the search
- To edit the search in the Query Translation box, add or delete terms and then click Search.
- Click URL to display the current search as a URL to bookmark for future use. Searches created using History numbers can not be saved using the URL feature.
- You may also save your search using My NCBI.
- The Result number link retrieves the documents found and displays them in a search results page.
- Translations details how each term was translated using Entrez's search rules and syntax for the database.
- User Query shows the search terms as you entered them in the search box and any syntax errors with the query.
|
 Search #1:
Search #1:
human arachidonic acid
will retrieve biosystems with those terms anywhere in the record.
Some of the records might be from organisms other than human because we did not limit that search term to the Organism search field. Therefore, we will also retrieve other species biosystems if they happen to contain the term "human" in a comment or some other field of the record.
Similarly, the term "arachidonic acid" can appear anywhere in the record. Some biosystems might have it in the title, and others might have it as a biosystem component.
The terms entered in a basic search may or may not be adjacent to each other in the retrieved records, depending on how Entrez parsed the query (as shown in the Search Details for a given search). To force terms to be searched as a phrase, use quotes. To refine your search in other ways, use the Limits option or the Advanced Search methods described below. |
Limits |
The Limits page allows you to restrict your search in various ways.
At a minimum, the Limits page displays the list of available search fields. You can do a separate search for each term or phrase in your query, as shown in sample Search #2 and #3 to the right, and select the desired search field for each one. (If desired, you can then combine the searches by using the Search Builder or History section of the Advanced Search page.)
For some databases, the Limits page also provides other commonly used options, as check boxes and/or pull-down menus, for restricting your search results to records with specific characteristics. These check boxes and pull-down menus generally represent a commonly used subset of the choices that are available from the Advanced Search page and are placed on the Limits page for easy access.
IMPORTANT NOTE: Once you have used a particular Limit, warning sign will appear near the top of your search results page that indicates which Limit(s) are currently in effect, for example:

Note that the Limit will remain in effect for all subsequent searches in the current database unless you change or remove that limit. In the illustrated example above, any search you do will be limited to the Titles of records, until you remove the limit.
|
 Search #2:
Search #2:
On the BioSystems search page, click on the Limits link, select the Organism search field, and enter the following query:
human
and press "GO". That will retrieve only biosystems found in human.
Search #3:
Open the Limits page again and clear your previous search. Change the search field selection to Title, enter the following query:
arachidonic acid
and press "GO". That will retrieve only records containing those terms in the title, or name, of a biosystem.
If desired, you can then combine the searches on the Advanced Search page, either by using the Search Builder, as shown in sample Search #4, or by using the History section of that page, as shown in sample Search #5.
|
| Advanced Search |
The Advanced Search page allows you to exercise greater control over your search, for example, by enabling you to:
- Build a search one step at a time.
- Browse the index of any search field and add term(s) of interest from the index to the active query box at the top of the page.
- View your search History and combine or subtract searches from each other.
As you build a query, either by using the Search Builder's pull-down menus, or by using the "Add" links in the "History" portion of the page to combine previous searches, the grey text box at the top of the page will display your current query.
You can also manually edit the current query by clicking the "Edit" link beneath the grey text box. That will allow you to type terms/search numbers/etc. directly into the box, add parentheses for nesting if desired, change Boolean operators, etc.
In addition, the following types of advanced searches can be entered in the query box of any Entrez search page (i.e., in the query box of the database's Home page, Limits page, or Advanced Search page):
(A companion tool, FLink, is available for Batch queries.)
|
Search Builder |
The "Search Builder" section of the Advanced Search page allows you to build your query step by step, adding a new search term and selecting a new search field at each step. It also allows you to browse the index of any search field to view the available terms.
To build a query:
(1) Select the Search Field of interest using the pull-down menu.
(2) Type a term(s) in the text box beside the search field menu. Or, use the "Show index list" link to see the index of the search field and select the desired term from the index. (tips on using the "Show Index List")
(3) Select the Boolean operator (AND, NOT, OR) that should precede the term when it is added to the active query at the top of the page.
Continue the above steps, as desired, to add more term/search field combinations to your query.
As you use the Search Builder, the grey text box at the top of the page will show your current query.
You can manually edit the current query by clicking the "Edit" link beneath the grey text box. That will allow you to type terms/search numbers/etc. directly into the box, add parentheses for nesting if desired, change Boolean operators, etc.
Press the Search button to display the records retrieved by your search (i.e., it displays the search results page).
Click on the "Add to history" link if you prefer to simply add the query to your search history and remain on the Advanced Search page, where you can continue building your query.
Tips on using the "Show Index List" function on the Advanced Search page:
The "Show Index List" function allows you to browse the index of any Search Field. If you select a search field and press the "Show Index" link without entering a term in the box, you will be taken to the top of the index. If you enter a term first, you will be taken to the part of the index that contains your term (or the closest alphabetical location, if your term is not present in the index).
The number of records that contain the term will appear in parentheses. You can also browse the index to explore the variety of terms available (for example, select "All Fields", enter "Huntington", and click on the "Show Index" link to see additional spellings and/or related terms, such as Huntington disease, Huntington's, Huntington's disease).
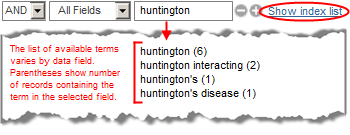
To select a range of terms from the index, use the Shift key while selecting the first and last term. Then use the AND, OR, or NOT buttons to add that group of terms to the active query.
To select multiple terms that do not fall within a continuous range from the index, use the Control key while selecting the terms of interest. Then use the AND, OR, or NOT buttons to add that group of terms to the active query.
Note: When multiple terms are selected from the index window, they are OR'ed together within parentheses and then appended to your query with whatever Boolean operator you have selected.
|
 Search #4:
Search #4:
On the BioSystems search page, click on Advanced Search and build your search one step at a time:
(a) Using the first pull-down menu in Search Builder, select the Organism search field and enter the following query:
human
and select "AND" as the Boolean operator. That term/search field selection will automatically be displayed in the grey text box at the top of the page, which shows your current query.
(b) Using the second pull-down menu in Search Builder, select the Title search field and enter the following query:
arachidonic acid
and select "AND" as the Boolean operator. That newest term/search field selection will automatically be added to the grey text box at the top of the page
(c) your query will now appear as:
human[Organism] AND arachidonic acid[Title]
Press the Search button if you want to display the records retrieved by your search (i.e., it displays the search results page).
Or, click on the "Add to history" link if you prefer to just add the query to your search history and remain on the Advanced Search page, where you can continue building your query.
Note that this search will produce the same results as sample searches #5 and #6. It is simply executed in a different way. That is, you remain on a single query page (Advanced search) and can browse the index of any search field as you build your query one step at a time.
|
History |
The "History" section of the Advanced Search page displays the searches you have done in the current database.
You can combine or subtract searches from each other by entering the search numbers and the AND, OR, or NOT Boolean operators in the query box, for example: #2 AND #3. If the query contains several search numbers and Boolean operators, the Boolean operators are processed from left to right unless parentheses are used for nesting. If parentheses are used, the portions of the query in parentheses will be processed first, then the remaining Boolean operators will be processed from left to right.
Additional details about Search History:
- The Search History will be lost after 8 hours of inactivity. (To save a search indefinitely, click on the search # and select "Save in My NCBI.")
- Click "Clear History" to delete all searches from History.
- Entrez will move a search statement number to the top of the History if a new search is the same as a previous search.
- History search numbers may not be continuous because some numbers are assigned to intermediate processes, such as displaying a citation in another format.
- The maximum number of searches held in History is 100. Once the maximum number is reached, PubMed will remove the oldest search from the History to add the most current search.
- A separate Search History will be kept for each database, although the search statement numbers will be assigned sequentially for all databases.
- PubMed uses cookies to keep a history of your searches. For you to use this feature, your Web browser must be set to accept cookies.
- Database records that you have copied to the Clipboard are represented by the search number #0, which may be used in Boolean search statements. For example, to limit the records you have collected in the Clipboard to those from human, use the following search: #0 AND human[organism]. This does not change or replace the Clipboard contents.
|
 Search #5:
Search #5:
Use the search numbers shown in the "History section" of the advanced search page to combine previous searches (for example, searches #2 and #3 shown above).
To do that, you can either:
Click on the "Edit" link beneath the grey text box and type in a search statement such as:
#2 AND #3
Or, instead of typing the search statement, use the "Add" link beside any search number in the "History" section of the Advanced Search page to add that search number into the grey text box.
That will retrieve only records that contain "human" in the Organism field and "arachidonic acid" in the Title field. Compare the retrieval from this search with that of the sample basic search above.
(Note that your search numbers might be different from those shown here, if you did earlier searches in the Entrez system before trying these examples.)
|
| Complex Boolean |
Whether you are on the Basic search page (i.e., the database's home page), the Limits page, or the Advanced search page, you can:
Enter a search in command language, specifying your exact combination of desired search terms, search fields, and Boolean operators, as shown in the examples to the right. The syntax is:
term[field] BOOLEAN term[field] BOOLEAN term[field] etc.
Search Field names must be placed in square brackets [], and can be written as either the full name, for example, [BioSystemType], or as the corresponding search field abbreviation, for example, [Type]. (additional examples)
Boolean operators (AND, OR, NOT) must be written in UPPER CASE.
Boolean operators are processed from left to right unless parentheses are used for nesting. If parentheses are used, the portions of the query in parentheses will be processed first, then the remaining Boolean operators will be processed from left to right.
Boolean operators can also be used to combine or subtract searches from each other (i.e., to find the union, difference, or intersection of the data sets retrieved by various searches). To do this, use the Search History section of the Advanced Search page and simply enter the search numbers and desired Boolean operators in the query box.
For example, to identify the records that were retrieved by Search #2 of your search history, and also by Search #3, you could enter the following query:
#2 AND #3
To identify the records that were retrieved by Search #2 but not by Search #3, you could enter the following query:
#2 NOT #3
|
 Search #6:
Search #6:
Simply enter all search terms and search fields as a single statement into the query box:
human[Organism] AND arachidonic acid[Title]
Note that this search will produce the same results as sample searches #4 and #5, but it takes only a single step when entered directly into the search box as a Boolean query.
Search #7:
(PTGS1[GeneName] OR "prostaglandin endoperoxide synthase 1"[GeneName]) NOT (primates[Organism] OR rodents[Organism])
This search will retrieve biosystem records that contain PTGS1 or prostaglandin endoperoxide synthase 1 in the GeneName field, but that will not be from any organisms in the taxonomic orders Primata or Rodentia.
|
| Range Search |
Range queries are constructed by specifying a lower and upper numerical value separated by a colon (:) to specify the range, followed by a search field name or abbreviation in square brackets, as shown in the examples to the right. You can insert a space on each side of the colon but that is not necessary; the search will work either way.
All "count" fields (such as CID count, SID count, Gene count, and Protein count) and all dates can be range queried.
Range queries on "counts" have the format:
FromCount : ToCount [fieldname]
Note: The FromCount and ToCount values are integers. An example is shown to the right, as Search #8. The search fields summary table includes the names and abbreviations for the various "counts" fields.
Range queries on Dates (such as CreateDate or ModifyDate) have a similar format:
FromDate : ToDate [fieldname]
Note: The FromDate and ToDate values can specify an exact date, a month, or a year, and are written in the format: YYYY/MM/DD, YYYY/MM, or YYYY. An example is shown to the right, as Search #9. The search fields summary table includes the names and abbreviations for the various "date" fields.
|
 Search #8:
Search #8:
8:15[GeneCount]
will retrieve biosystems that are linked to as few as 8 Gene IDs through as many as 15 Gene IDs.
Search #9:
2011/04/10[CDAT] : 2011/05/25[CDAT]
will retrieve records that have a CreateDate [CDAT] from April 10, 2011 through May 25, 2011 (i.e., it will retrieve records that were first imported into the NCBI BioSystems database between those dates).
|
| Batch Query |
 FLink is a tool that was developed to handle large quantities of input and output data. It enables you to traverse from a group of records in a source database to a ranked list of associated records in a destination database. FLink is a tool that was developed to handle large quantities of input and output data. It enables you to traverse from a group of records in a source database to a ranked list of associated records in a destination database.
For example, it can accept up to 100,000 protein sequence, gene, or small molecule identifiers as input and retrieve a ranked list of up to 100,000 biosystems.
An illustrated quick start guide shows the easy 1-2-3 step process for using the tool. The help document also provides details about the proper formatting of the (1) input UID list, the features of the (2) review + select display and (3) output display.
Although  FLink was initially developed as a companion tool for the BioSystems database, it can also be used in a similar way for other types of input and output data, as noted in the list of FLink's supported databases. FLink was initially developed as a companion tool for the BioSystems database, it can also be used in a similar way for other types of input and output data, as noted in the list of FLink's supported databases.
|

|
Additional details about search methods and options are provided in the: (1) PubMed help document (including information about temporarily saving records from your search results to the Clipboard); (2) My NCBI help document (including information about Saving search strategies and indefinitely saving records from your search results into your My NCBI Collections); and (3) general Entrez help document.
 
By default, Entrez searches All Fields of the database unless a specific search field is indicated in the query. Search fields can be selected from pop-up menus on either the Limits and Advanced Search page, or can be typed directly in your query (surrounding field names with square brackets [], for example, [Organism] or [Orgn]).* The Show Index link on the Advanced Search page allows you to browse the index of each search field, where you can see the available terms, the number of records containing each term or phrase, as well as the syntax for entering values in search fields such as CreateDate or ModifyDate.
The currently available fields include:
| Field name |
Abbreviation* |
Description |
Sample Search |
| All Fields |
[ALL] |
Searches the complete database record |
 "arachidonic acid"[all]
"arachidonic acid"[all]
will retrieve the Biosystems records that contain the phrase "arachidonic acid" in any field of the record.
(Compare these search results with those obtained by the sample Title field search, which will retrieve records containing that phrase only in the biosystem title.)
The quotes surrounding the search terms ensure they are searched as a phrase. |
| Accession |
[ACCN] |
Searches only the accession number of the biosystem record, which is always an alphanumeric combination (e.g., bsid12345).
(If you enter only the digits from the accession (e.g., 12345), use the UID field instead) |
 bsid82991[accn]
bsid82991[accn]
will retrieve the biosystem record that contains the specified unique identifier in the accession number field. |
| BioSystemType |
[Type] |
Allows you to retrieve, or limit your search results to, a specific type of biosystem record. Currently available types include:
| pathway |
A series of related biochemical reactions. Some source databases, such as KEGG and Reactome, organize pathways hierarchically into supersets and subsets (KEGG refers to its subsets as "pathway modules"). The BioSystems database includes both supersets and subsets and provides cross-references between them. This category of records in the NCBI BioSystems database also includes biological processes from Gene Ontology (GO).
 |
|
| structural complex |
Biomolecules and/or chemicals that are bound to each other as asserted by the source database. This category of records in the NCBI BioSystems database also includes cellular components from Gene Ontology (GO). |
|
| signature module |
A group of genes that have a pathogenicity or other phenotype associated with them. |
|
| functional set |
A group of enzymes that perform a certain function, for example, all aminoacyl transferases. This category of records in the NCBI BioSystems database also includes molecular functions from Gene Ontology (GO). |
|
|
 pathway[BioSystemType]
pathway[BioSystemType]
OR
pathway[Type]
will retrieve only pathways from the the BioSystems database.
Similarly:
structural complex[Type]
will retrieve all structural complex records from the NCBI BioSystems database, and
functional set[Type]
will retrieve all functional set records from the NCBI BioSystems database.
|
| CID |
[CID] |
PubChem Compound identifiers (CIDs) found in a biosystem using the method descibed in the data processing/create direct links/small molecules section of this document. |
 5280883[cid]
5280883[cid]
will retrieve biosystems that include prostaglandin G2 (PGG2, CID 5280883) as a component. |
| CIDCount |
[CIDCount] |
Total number of PubChem Compound identifiers (CIDs) linked to a biosystem using the method descibed in the data processing section of this document.
Fields such as CID count, SID count, Gene count, and Protein count, taken individually or together, give an indication of the relative size of the biosystem record. For example, BioSystem records with large counts may be Superset biosystems, whereas records with small counts may be Subset biosystems. |
 10:23[CIDCount]
10:23[CIDCount]
will retrieve biosystems that are linked to as few as 10 PubChem CIDs through as many as 23 PubChem CIDs.
(more about range searching...) |
| ChemicalName |
[CN] |
Names of small molecules that are components in a biosystem. The names include the first four synonyms listed in the PubChem compound records that have been associated with the biosystem through the method described in data processing/create direct links/small molecules. For example, the first four synonyms listed in PubChem CID 5280883 are prostaglandin G2, PGG2, CHEBI:27647, and LMFA03010009; a search for any one of these names will retrieve biosystems that include PGG2 as a component. |
 PGG2[cn]
PGG2[cn]
will retrieve biosystems that include PGG2 as a component. |
| Comments |
[COM] |
Comments and additional information provided by the source database on the biosystem |

|
| CreateDate |
[CDAT]
[PDAT]
[DP] |
The date the biosystem record first appeared in the NCBI biosystems database. This is sometimes referred to as the publication date, hence the synonymous abbreviations PDAT and DP. The syntax for searching the field is YYYY/MM/DD, YYYY/MM, or YYYY. The colon (:) can be used to search for a range of dates, for example, YYYY/MM/DD:YYYY/MM/DD[CDAT]. |
 2011/04/10[CDAT] : 2011/05/25[CDAT]
2011/04/10[CDAT] : 2011/05/25[CDAT]
will retrieve records that were first imported into the NCBI BioSystems database from April 10, 2011 through May 25, 2011.
(more about range searching...)
|
| Description |
[DESC] |
The text description of a biosystem provided by the source database. |

|
| Filter |
[FILT] |
The "Filter" search field allows you to narrow your retrieval to records that have certain attributes, such as record type (e.g., conserved biosystem or organism specific biosystem).
It also allows you to limit search results to biosystem records that have links to other Entrez databases of interest, as shown in the sample search to the right. A detailed explanation of each type of link is provided in the description of an Entrez search results page.
The Filter field can also be used to view current database statistics, by entering a search for All[Filt], as shown in the example in the next column. |
 conserved biosystem[filt]
conserved biosystem[filt]
will retrieve only that record type from the BioSystems database.
biosystems_structure[filt]
will retrieve the BioSystems records that have associated data in the Entrez Structure database.
You can then open the "Display" menu near the top of the BioSystems search results page and select Structures to retrieve the corresponding Entrez Structure records (for all biosystems records you have retrieved, or only for those whose checkboxes have been activated). The 3D structure records can then be viewed interactively using the free Cn3D software program.
all[filt]
will retrieve all of the biosystem database records, showing the total number retrieved.
|
| GeneCount |
[GeneCount] |
Total number of Entrez Gene IDs linked to a biosystem using the method descibed in the data processing section of this document.
Fields such as CID count, SID count, Gene count, and Protein count, taken individually or together, give an indication of the relative size of the biosystem record. For example, BioSystem records with large counts may be Superset biosystems, whereas records with small counts may be Subset biosystems. |
 8:15[GeneCount]
8:15[GeneCount]
will retrieve biosystems that are linked to as few as 8 Gene IDs through as many as 15 Gene IDs.
(more about range searching...) |
| GeneExternalID |
[GEID] |
The identifier given to a gene by the source of a biosystem. |

|
| GeneID |
[GID] |
NCBI Entrez Gene IDs found in a biosystem using the method descibed in the data processing/create direct links/genes section of this document. Entrez GeneIDs are numerical identifiers for a genetic locus that remain stable regardless of changes to the locus name, coordinates, or sequence data. |
 5742[gid]
5742[gid]
will retrieve biosystems that include the Entrez GeneID 5742 (human PTGS1 gene) as a component. |
| GeneName |
[GN] |
Gene name, including all of the synonyms found in the corresponding Entrez Gene record. For example, prostaglandin-endoperoxide synthase 1 (GeneID 5742) has the following synonyms listed in its Entrez Gene record: COX1; COX3; PHS1; PCOX1; PGHS1; PTGHS; PGG/HS; PGHS-1; PTGS1. Any of these synonyms can be searched in the Gene Name field to retrieve biosystems that include the gene as a component. |
 PTGS1[gn]
PTGS1[gn]
will retrieve biosystems that include the human PTGS1 gene (Entrez GeneID 5742) as a component. |
| ModifyDate |
[MDAT] |
The date the biosystem record last changed in the NCBI biosystems database. The syntax for searching the field is YYYY/MM/DD, YYYY/MM, or YYYY. The colon (:) can be used to search for a range of dates, for example, YYYY/MM/DD:YYYY/MM/DD[MDAT]. |
 2011/04/10[MDAT] : 2011/05/25[MDAT]
2011/04/10[MDAT] : 2011/05/25[MDAT]
will retrieve biosystem records that were modified from April 10, 2011 through May 25, 2011.
(more about range searching...)
|
| Organism |
[ORGN] |
Organism(s) in which a biosystem is found. A common name (e.g., human), scientific name (e.g., Homo sapiens), or other taxonomic node (e.g, Primates or Primata) can be entered as a query.
Note that a search of the organism field will retrieve only organism specific biosystems that fall under the specified taxonomic node. To find corresponding conserved biosystems that are present in numerous organisms, simply follow the "related biosystems/conserved biosystems" link from the biosystem record(s) of interest. |
 human[orgn]
human[orgn]
will retrieve biosystems found in human.
primates[orgn]
will retrieve biosystems found in individual species falling in the order Primata.
|
| ProteinCount |
[ProteinCount] |
Total number of Entrez Protein IDs linked to a biosystem using the method descibed in the data processing section of this document.
Fields such as CID count, SID count, Gene count, and Protein count, taken individually or together, give an indication of the relative size of the biosystem record. For example, BioSystem records with large counts may be Superset biosystems, whereas records with small counts may be Subset biosystems. |
 15:28[ProteinCount]
15:28[ProteinCount]
will retrieve biosystems that are linked to as few as 15 Protein IDs through as many as 28 Protein IDs.
(more about range searching...) |
| ProteinID |
[PID] |
This field contains the GI numbers and accession numbers of proteins found in a biosystem using the method descibed in the data processing/create direct links/proteins section of this document. |
 A search for either
A search for either
AAA03630[pid]
OR
189887[pid]
will retrieve biosystems containing the protein sequence record that has accession number AAA03630 (GI number 189887).
|
| ProteinName |
[PN] |
Names of proteins in a biosystem. The names are extracted from the definition line of protein sequence records that have been associated with the biosystem using the method descibed in the data processing/create direct links/proteins section of this document. |
 "prostaglandin endoperoxide synthase 1"[pn]
"prostaglandin endoperoxide synthase 1"[pn]
will retrieve biosystems containing a protein component with that name.
The quotes surrounding the search terms ensure they are searched as a phrase. |
| SID |
[SID] |
PubChem Substance identifiers (SIDs) found in a biosystem using the method descibed in the data processing/create direct links/small molecules section of this document. |

|
| SIDCount |
[SIDCount] |
Total number of PubChem Substance IDs linked to a biosystem using the method descibed in the data processing section of this document.
Fields such as CID count, SID count, Gene count, and Protein count, taken individually or together, give an indication of the relative size of the biosystem record. For example, BioSystem records with large counts may be Superset biosystems, whereas records with small counts may be Subset biosystems. |
 12:25[SIDCount]
12:25[SIDCount]
will retrieve biosystems that are linked to as few as 12 Protein IDs through as many as 25 PubChem SIDs.
(more about range searching...) |
| SidExternalID |
[SEID] |
External registry names for small molecules that are found in a biosystem record, using the method descibed in the data processing/create direct links/small molecules section of this document. |

|
| SourceAccession |
[SACC] |
The accession number assigned to a biosystem by the source database.
For example, the NCBI BioSystem record bsid82991, for human arachidonic acid metabolism, is derived from KEGG record hsa00590. Hsa00590 is the source accession. |
 hsa00590[sacc]
hsa00590[sacc]
will retrieve the KEGG organism specific biosystem for human arachidonic acid metabolism.
ko00590[sacc]
will retrieve the KEGG conserved biosystem for arachidonic acid metabolism. |
| SourceID |
[SRCID]
[SRID] |
A numerical identifier that is assigned to a particular source database of biosystem records. |
 1[srcid]
1[srcid]
will retrieve records deposited by the BioCyc database.
2[srcid]
will retrieve records deposited by the KEGG database.
|
| SourceName |
[SRC]
[SRCN]
[SRNM] |
Name of the organization that is the source of the record. (See the source databases section of this document for details.)
You can also see the allowable values in the SourceName field by using the Show index link on the Advanced Search page. |
 biocyc[src]
biocyc[src]
will retrieve records deposited by the BioCyc database.
kegg[src]
will retrieve records deposited by the KEGG database.
|
| Title |
[TI]
[TITL] |
Contains the brief title (or name) of the biosystem, for example, "arachidonic acid metabolism", or "citrate cycle", or "TCA cycle", etc. as assigned by the source database. |
 "arachidonic acid"[ti]
"arachidonic acid"[ti]
will retrieve records with that phrase in the biosystem title.
(Compare these search results with those obtained by the sample All Fields search, which will retrieve biosystem records containing that phrase anywhere in the record.)
The quotes surrounding the search terms ensure they are searched as a phrase. |
| UID |
[UID] |
Retrieves a biosystem record by its unique identification number, which is the numerical portion of the Accession number. For example, if an accession number is bsid12345, the corresponding UID is 12345.
If you enter a string of digits as a query and do not specify a search field, the UID field will be searched by default. |
 82991[UID]
82991[UID]
will retrieve the biosystem record whose unique identification number is 82991.
82991
will also retrieve that same biosystem record, because the UID field is searched by default for queries that are only a string of digits.
|
* In a query, the field name may be typed as the full name or abbreviation, and may be in upper, lower, or mixed case. If more than one abbreviation is shown, any one of them can be used. The field name must be surrounded by square brackets []. A space between the search term and the field specifier is optional. If desired, surround a phrase with quotes to force an adjacency search. For example, the sample queries below will work equally:
"arachidonic acid"[TI]
"arachidonic acid"[TITL]
"arachidonic acid" [TITL]
"arachidonic acid" [titl]
"arachidonic acid"[Title]
** The quotes surrounding the query terms in some of the sample searches force the terms to be searched as a phrase. If quotes are not used, the Entrez system may still recognize and handle the terms as a phrase, if they are present in a phrase dictionary used by the search engine. If the terms are not present in the phrase dictionary and are not surrounded by quotes, Entrez will insert a Boolean AND between the terms; in that case, they may or may not appear adjacent to each other in the retrieved records. The "Details" folder tab on a search results page will show you exactly how the Entrez system parsed your query. More search tips are provided in the PubMed help document and Entrez help document.
It is also possible to search for a word stem by using an asterisk (*) as a wild card; for example, arachidon* will retrieve records with terms such as arachidonate, arachidonic, arachidonoyl. The Entrez Help document provides additional information about truncating search terms in this way.
 
The Entrez databases to which biosystems records have been linked (via the data processing pipeline) generally have reciprocal links from their records back to the corresponding NCBI BioSystems database records.
Therefore, if you start your search in an Entrez database other than NCBI BioSystems, you can view the "Links" menus of the records retrieved to see if they have links to associated information in the NCBI BioSystems database. Alternatively, you can use the "Find related data" menu in the right hand margin of an Entrez search results page (in whatever database you have chosen to search) and select "BioSystems" to view the associated biosystem records for all items (default) displayed on the search results page or for those you have selected using their checkboxes.
For example:
- retrieve a record for a gene or protein of interest from the Entrez Gene or Entrez Protein database, such as Gene record 5742: human PTGS1, or the protein sequence record for its gene product, NP_000953: prostaglandin G/H synthase 1 isoform 1 precursor [Homo sapiens].
- view the "Links" menu in the right margin of the Gene record, or the "Related information" menu in the right margin of the Protein record, and select "BioSystems."
- If you are looking at a Gene record, the "General Gene Information: Pathways from BioSystems" section of the record provides another way to view/link to the associated biosystems. (As an example, see GeneID 5742: human PTGS1: general gene information.)
- Click on a biosystem of interest to view its record. The gene record from which you started will appear at the top of the "genes" folder tab for that biosystem and will be selected by default (i.e., its check box will be activated). You can then click on the "Highlight selected records in source database" to view the location of the gene within the pathway diagram, if that function is available for a given biosystem. (Currently, the highlight function is available only for KEGG records and the selected gene will be displayed with a red outline, as shown in an illustrated example. The gene name will also appear in red font, as may other gene names in the pathway diagram, indicating a link to the corresponding KEGG gene record. The red outline, however, will only appear around the gene(s) that were selected in the NCBI biosystem record.)
- The biosystems that are retrieved have been associated with the gene or protein through the methods descibed in the data processing/create direct links section of the BioSystems help document.
|
|
|
|
|
|
 |
|
|
|
|
| document summary page | display settings: format, items per page, sort by | send to | filter your results | refine your results | find related data |
 
The initial search results provide a list (document summary, or "DocSum") of the biosystem records that contain your search term, which can appear in any field of the record, unless a search field was specified in the query.
If desired, you can narrow your search by restricting the query to a search field of interest or adding more terms with a Boolean AND.
Alternatively, you can broaden your search by adding more terms (e.g., synonyms) to your query with a Boolean OR, or by following links to Related BioSystems.
Once you are satisfied with your search results, click on the BSID of any record on the DocSum page to view its biosystem summary page. In addition, the following options are available for viewing the search results:
 
The "Display settings" menu on acts upon all of the structure records (default) in your search results, or on the subset you have selected with checkboxes. You can select items from multiple pages of the search results, if desired.
| Format |
Summary *-- a summary of all of the structure records (default) retrieved by your search, or for those you have selected with checkboxes, in HTML format.The information shown for each record may include the following, as available:
- Biosystem title (biosystem name). You can click on the title of any biosystem to view its summary page.
- The first 250 characters of the description, if one has been provided by the source database
- Record type (pathway, structural complex, signature module, or functional set)
- Taxonomic scope (organism specific biosystem or conserved biosystem)
- Organism in which the biosystem is found (no organism name is displayed for conserved biosystems)
- BSID
- Source database and corresponding source accession
- Links to the genes, proteins, and small molecules that are associated with the biosystem. The associations are made using the methods described in the data processing section of this help document. The genes, proteins, and small molecules are also accessible from folder tabs in the biosystem's summary page. (Note: The "Find Related Data" menu in the right margin of the search results page provides a complete list of links to data associated with the biosystems. That menu retrieves related data for all records (default) retrieved by your search, or for the subset of records you have selected with checkboxes.)
Summary (text) -- a summary of the records retrieved by your search, in plain text format. The information shown for each record is the same as in the "Summary" format described above, but does not include the links to the genes, proteins, and small molecules. This setting displays all of the structure records (default) retrieved by your search, or those you have selected with checkboxes.
Abstract *-- similar to "Summary" format described above, with the difference that "Abstract" format displays the complete description, if one is available, rather than the first 250 characters. This setting displays all of the structure records (default) retrieved by your search, or those you have selected with checkboxes.
UI List -- a list of the unique identifiers (UI's) for all of the biosystem records (default) retrieved by your search, or for those you have selected with checkboxes.
|
| Items per page |
By default, 20 documents are listed per page. If desired, decrease (to a minimum of 5) or increase (to a maximum of 200) the number of documents displayed per page then press the "Apply" button.
|
| Sort by |
Search results are displayed in order of decreasing relevance with respect to the query. Many search fields have a score or rank associated with them; for example, the Title and Organism fields have a high rank, while the Comments field has a lower rank. The presence of a search term in any one or more of the fields is scored accordingly by the search system, and the total score given to a hit is used in determining its relevance to the query and therefore its placement on the search results page.
Currently, the only other sort option is to display by descending or ascending order of modification date.
Technical note: If you retrieve all records in the database by searching the Filter field for All[Filt], the records are simply displayed in descending order of BSID.
|
* The "Summary" and "Abstract" formats might appear very similar to each other if a search retrieves primarily biosystems that do not have descriptions. The variation in display formats can be easily seen, however, if the search results includes biosystems that have descriptions, such as the records retrieved by a search for human TCA cycle. Regardless of which display format you choose, you can click on the title of any biosystem to view its summary page.
 
The "Send To" menu options act upon all the hits retrieved by your search (default), or those you have selected by using their checkboxes.
| File |
Saves all the hits retrieved by your search into a plain text file, in either "Summary (text)" or "UI List" format.
|
| Clipboard |
Copies all the hits retrieved by your search (default), or those you have selected with check boxes, into a Clipboard, which temporarily stores up to 500 items (they will be lost after 8 hours of inactivity).
Click on the "Clipboard: XX items" link in the upper right corner of the page to view the items in any format for up to 8 hours after your last activity in the database.
The Clipboard will not add an item that is currently in the Clipboard; it will not create duplicate entries. You can remove items from the Clipboard, if desired.
Entrez uses cookies to add your selections to the Clipboard. For you to use this feature, your Web browser must be set to accept cookies.
Items in the Clipboard are represented by the search number #0, which may be used in Boolean search statements. For example, to limit the items you have collected in the Clipboard to those from human, use the following search: #0 AND human[organism]. This does not affect or replace the Clipboard contents.
The Clipboard's "Send to" menu offers you the same "File" and "Collections" options as offered on the original search results page. The latter option saves all items (default), or the subset of items selected with check boxes, indefinitely in the My NCBI Collections section of your My NCBI account.
|
| Collections |
Saves all the hits retrieved by your search (default), or those you have selected by using their checkboxes, into the My NCBI Collections section of your My NCBI account.
|
 
The "Filter your results" area in the upper right corner of a search results page allows you to see all the records (default) retrieved by your search, or subsets of your search results that reflect commonly requested categories of records, and shows the corresponding number of records in each case.
Specifically, the "Conserved BioSystems" and "Organism Specific BioSystems" links show the number of records in your search results that fall into those record categories, and enable you to view those subsets of biosystems, if desired. The Refine your results box enables you to view other subsets from your search results.
|
 

The "Refine your results" panel that appears in the right hand margin of a search results page allows you to retrieve the subsets of biosystem records from your search results that: (1) fall into specific record categories; (2) come from specific source databases; or (3) include proteins and compounds that have corresponding bioactivity data in the PubChem BioAssay database.
The value in the parentheses indicates the number of biosystems from your search results that fall into each listed category. Click on any link in the "Refine your results" panel to retrieve the corresponding subset of biosystems:
- Record Type:
Pathway - The subset of biosystems in your search results that contain series of related biochemical reactions. The "pathway" category of records in the NCBI BioSystems database also includes "biological processes" from Gene Ontology (GO).
Structural Complex - The subset of biosystems in your search results that contain biomolecules and/or chemicals that are bound to each other, as asserted by the source database. The "structural complex" category of records in the NCBI BioSystems database also includes "cellular components" from Gene Ontology (GO).
Functional Set - The subset of biosystems in your search results that contain groups of enzymes that perform a certain function, for example, all aminoacyl transferases. The "functional set" category of records in the NCBI BioSystems database also includes "molecular functions" from Gene Ontology (GO).
Signature Module - The subset of biosystems in your search results that contain a group of genes that have a pathogenicity or other phenotype associated with them.
Additional details about each category are provided in the help section on "Content of the BioSystems Database: Record Types."
- Source Name:
BioCyc - The subset of biosystems in your search results that are from BioCyc (http://biocyc.org/), a collection of organism-specific pathway/genome databases (PGDBs). The Tier 1 (EcoCyc (http://ecocyc.org/) and MetaCyc (http://metacyc.org/)) and Tier 2 data sets are included in the NCBI BioSystems database.
Gene Ontology (GO) - The subset of biosystems in your search results that are from the Gene Ontology (GO) project (http://www.geneontology.org/), an initiative to standardize the representation of gene and gene product attributes across species and databases and provides a controlled vocabulary of terms for describing gene product characteristics and gene product annotation data..
KEGG - The subset of biosystems in your search results that are from KEGG: Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/) by the Kanehisa Laboratory of the Bioinformatics Center, Institute for Chemical Research, Kyoto University, Kyoto, Japan.
LIPID MAPS - The subset of biosystems in your search results that are from LIPID Metabolites And Pathways Strategy (LIPID MAPS, http://www.lipidmaps.org/), a multi-institutional effort to identify and quantitate, using a systems biology approach and sophisticated mass spectrometers, the lipid species in mammalian cells, and to quantitate the changes in these species in response to perturbation.
Pathway Interaction Database (PID) - The subset of biosystems in your search results that are from Pathway Interaction Database (PID, http://pid.nci.nih.gov/) "a freely available collection of curated and peer-reviewed pathways composed of human molecular signaling and regulatory events and key cellular processes."
Reactome - The subset of biosystems in your search results that are from Reactome (http://www.reactome.org/), a curated knowledge base of human biological pathways and inferred equivalent reactions in multiple non-human species.
WikiPathways - The subset of biosystems in your search results that are from WikiPathways (http://www.wikipathways.org/index.php/WikiPathways), an open, public platform dedicated to the curation of biological pathways by and for the scientific community.
Additional details about each source are provided in the help section on "Content of the BioSystems Database: Source Databases."
- BioAssays:
|
 
 As noted in the page on discovering associations among previously disparate data, the Entrez retrieval system is designed to provide integrated access to previously disparate data and make it possible to collect related information on a topic of interest within and across Entrez databases.
As noted in the page on discovering associations among previously disparate data, the Entrez retrieval system is designed to provide integrated access to previously disparate data and make it possible to collect related information on a topic of interest within and across Entrez databases.
As part of Entrez, the NCBI BioSystems database implements a data processing pipeline to identify such associations and present them as link options on search results pages.
The "Related information" box that appears in the right margin of the display for an individual record allows you to retrieve related data for that particular structure. For example, if you select "Conserved Domains" when you are viewing the record for accession 1PTH, you will retrieve the domain models from the Conserved Domain Database that have been annotated on the protein molecules in that structure. Many of the links are also available on a biosystem's summary page.
The data accessible through some of the Links (e.g., genes, proteins, small molecules) are also presented in folder tabs on the individual BioSystem records. The folder tabs may present the data in a different way and/or provide additional functions not available through the links menus. For example, the Proteins folder tab sorts identical proteins by PIG ID, and the Genes and Small Molecules folder tabs provide the ability to highlight selected components in the source database's full size diagram if that function is available at the source database. In addition, other small differences may exist, as noted in the descriptions of the Genes and Citations folder tabs.
A "Find Related Data" box (instead of an "Related information" box) will appear in the right margin of an Entrez Structure search results page if you retrieved two or more records. The "Find Related Data" box allows you to retrieve related data for all the records retrieved by your search (default), or for the records you have selected with checkboxes. For example, you can use this to view the list of genes/proteins/small molecules associated with all/selected biosystems. The related data will then be displayed in the default sort order for their database, such as by date or by UID.
(Tip: If you prefer to view the genes/proteins/small molecules as a ranked list that reflects how frequently each item was found in the biosystems retrieved by your search, use the FLink tool. The About FLink page provides an overview of the tool and the FLink help document provides details on how to use it. Note that you can use an Entrez Search as input for a FLink operation. For example, after you do you BioSystems search, open FLink, choose the BioSystems database to start, select the desired search from the "Input from Entrez History" folder tab and press "submit." That will open your BioSystems search result in the FLink tool, where you can now use the "LinkTo" menu to select the data type (genes, proteins, small molecules, etc.) that you want to view as a ranked list.)
|
| Link Group |
Link Name  |
Description |
| Sequences |
Genes |
 Genes involved in the biosystem. The association between genes and biosystems was made using the method descibed in the data processing/create direct links/genes section of this document.
Genes involved in the biosystem. The association between genes and biosystems was made using the method descibed in the data processing/create direct links/genes section of this document. |
| Proteins |
 Proteins involved in the biosystem. The association between proteins and biosystems was made using the method descibed in the data processing/create direct links/proteins section of this document.
Proteins involved in the biosystem. The association between proteins and biosystems was made using the method descibed in the data processing/create direct links/proteins section of this document. |
| Conserved Domains |
 Conserved domains that are specific hits to the proteins involved in biosystems, representing high confidence functional annotations. The association between proteins and biosystems was made using the
method descibed in data processing/create direct links/proteins.
Conserved domains that are specific hits to the proteins involved in biosystems, representing high confidence functional annotations. The association between proteins and biosystems was made using the
method descibed in data processing/create direct links/proteins. |
| HomoloGene |
 HomoloGene records that are linked to the proteins involved in the biosystem. The HomoloGene records list putative orthologs of the genes that coded the proteins.
HomoloGene records that are linked to the proteins involved in the biosystem. The HomoloGene records list putative orthologs of the genes that coded the proteins.
|
| ProteinClusters |
 Protein Clusters database records that are linked to the proteins involved in the biosystem. The Protein Clusters database contains both curated and uncurated clusters of proteins (from four major groups: prokaryotes, bacteriophages and the mitochondrial and chloroplast organelles) that are grouped by sequence similarity.
Protein Clusters database records that are linked to the proteins involved in the biosystem. The Protein Clusters database contains both curated and uncurated clusters of proteins (from four major groups: prokaryotes, bacteriophages and the mitochondrial and chloroplast organelles) that are grouped by sequence similarity.
|
| Small Molecules |
PubChem Compounds |
 Compounds involved in biosystems. The association between compounds and biosystems was made using the method descibed in the data processing/create direct links/small molecules section of this document.
Compounds involved in biosystems. The association between compounds and biosystems was made using the method descibed in the data processing/create direct links/small molecules section of this document.
|
| PubChem Substances |
 Substances involved in biosystems. The association between substances and biosystems was made using the method descibed in the data processing/create direct links/small molecules section of this document..
Substances involved in biosystems. The association between substances and biosystems was made using the method descibed in the data processing/create direct links/small molecules section of this document..
|
| Related BioSystems |
Organism Specific BioSystems |
 The organism specific biosystems that correspond to a conserved biosystem, as determined by the source database.
The organism specific biosystems that correspond to a conserved biosystem, as determined by the source database.
|
| Conserved BioSystems |
 The conserved biosystem, if one is available, that corresponds to an organism specific biosystem. Conserved biosystems are canonical biosystems that are used to group together orthologous, organism specific biosystems. They are derived from "reference pathways" in the KEGG database and from MetaCyc records.
The conserved biosystem, if one is available, that corresponds to an organism specific biosystem. Conserved biosystems are canonical biosystems that are used to group together orthologous, organism specific biosystems. They are derived from "reference pathways" in the KEGG database and from MetaCyc records. |
| Linked BioSystems |
 Biosystems that the source database has explicitly stated are connected in some way.
Biosystems that the source database has explicitly stated are connected in some way. |
| Similar BioSystems |
 Biosystems share at least one identical protein sequence from the same source organism. In other words, the shared protein is in the same protein identity group (PIG) and has the same Taxonomy ID (TaxID).
Biosystems share at least one identical protein sequence from the same source organism. In other words, the shared protein is in the same protein identity group (PIG) and has the same Taxonomy ID (TaxID). |
| Superset BioSystems |
 Biosystems that are one node above the one currently being viewed, according to a hierarchical organization used by the source database.
Biosystems that are one node above the one currently being viewed, according to a hierarchical organization used by the source database.
For example, "Phase 1 - Functionalization of compounds" (bsid105699) is a superset of "COX reactions" (bsid105711), reflecting the hierarchical organization of those biosystems specified by the source database, Reactome. |
| Subset BioSystems |
 Biosystems that are one node below the one currently being viewed, according to a hierarchical organization used by the source database.
Biosystems that are one node below the one currently being viewed, according to a hierarchical organization used by the source database.
For example, "COX reactions" (bsid105711) are a subset of "Phase 1 - Functionalization of compounds" (bsid105699), reflecting the hierarchical organization of those biosystems specified by the source database, Reactome. |
| Literature |
PubMed Citations |
 PubMed records whose PMIDs were explicitly cited in (and extracted from) the source database record.
PubMed records whose PMIDs were explicitly cited in (and extracted from) the source database record. |
| OMIM |
 Records from the Online Mendelian Inheritance in Man (OMIM) database that are linked to the Entrez Gene records associated with a BioSystem. The association between genes and biosystems was made using the method descibed in the data processing/create direct links/genes section of this document.
Records from the Online Mendelian Inheritance in Man (OMIM) database that are linked to the Entrez Gene records associated with a BioSystem. The association between genes and biosystems was made using the method descibed in the data processing/create direct links/genes section of this document. |
| Other Links |
BioAssays via Actives |
 BioAssays in which compounds from biosystems have been found to be active. The association between the compounds and biosystems was made using the method descibed in the data processing/create direct links/small molecules.
BioAssays in which compounds from biosystems have been found to be active. The association between the compounds and biosystems was made using the method descibed in the data processing/create direct links/small molecules.
Tip: If you would like to see whether a specific compound (CID) has been found to be active in a bioassay record, click on the CID of interest in the small molecules folder tab of a biosystem record to view the corresponding PubChem Compound record and see the various "Links" available for that record. |
| BioAssays via Target |
 BioAssays whose protein targets are involved in biosystems.
BioAssays whose protein targets are involved in biosystems.
The association between the proteins and biosystems was made using the method descibed in data processing/create direct links/proteins.
If the proteins, in turn, have their GI numbers cited as targets in PubChem BioAssay records, the BioAssays via Target link retrieves the subset of bioassay records in which those proteins are targets of an assay.
Tip: If you would like to see whether a specific protein has been used as the target of a bioassay, click on the GI number of interest in the proteins folder tab of a biosystem record to view the corresponding sequence record in the Entrez Protein database. The "Related information" menu in the right margin of the protein sequence record will indicate whether that protein was the target of any bioassays. If the menu includes an option for "BioAssay by Target", it means protein's GI number is specifically named as a target in a BioAssay record(s), and the BioAssay(s) can be retrieved by clicking on the link. If that GI was not specifically named as a target but an identical protein sequence (belonging to the same PIG) was named, the "Related information" menu will include an option for "BioAssay by Target, identical sequence." |
| Structures |
 3D macromolecular structures of proteins involved in biosystems.
3D macromolecular structures of proteins involved in biosystems.
The 3D protein structures are from the Molecular Modeling Database (MMDB), also known as "Entrez Structure," and can be viewed with Cn3D, a free helper application available for Windows, Macintosh, and Unix platforms. A tutorial describes the Cn3D program's features and functions.
Tip: If you would like to see whether a specific protein in a biosystem has a resolved 3D structure, click on the GI number of interest in the proteins folder tab of a biosystem record to view the corresponding sequence record in the Entrez Protein database. The "Related information" menu in the right margin of the protein sequence record will indicate whether a 3D structure is available, and what type of association exists between the sequence and structure:
an option for "Structure" will appear in the Links menu if the protein was derived directly from a 3D structure record
an option for "Related Structure" will appear in the Links menu if similar protein sequences, identified via BLAST, were derived from 3D structures
(The "Other Links:Structures" option in a BioSystem record retrieves the former subset of structures but not the latter.) |
| Taxonomy |
 This link retrieves the NCBI Taxonomy database record for the source organism whose Taxonomy ID (TaxID) was provided in the source record. For additional information, see the data processing/create direct links/taxonomy section of this document.
This link retrieves the NCBI Taxonomy database record for the source organism whose Taxonomy ID (TaxID) was provided in the source record. For additional information, see the data processing/create direct links/taxonomy section of this document. |
If the  FLink icon appears beside a link name, clicking on that icon will open the data as a ranked list in the "FLink" tool. Clicking on the link name, in contrast, will open the data directly in the corresponding database, in the default sort order for that database. The About FLink page provides an overview of the tool, and the FLink help document provides additional detail. FLink icon appears beside a link name, clicking on that icon will open the data as a ranked list in the "FLink" tool. Clicking on the link name, in contrast, will open the data directly in the corresponding database, in the default sort order for that database. The About FLink page provides an overview of the tool, and the FLink help document provides additional detail.
 
Regardless of the format in which you have chosen to display your search results, simply click on the BSID or biosystem title for a record of interest to view its BioSystem record.
|
|
|
|
|
|
 |
|
|
|
|
| |
| BIOSYSTEM RECORD: COMPONENTS AND FEATURES |
An individual biosystem record (for example, bsid82991) includes descriptive information, a thumbnail diagram, if available, and folder tabs that categorize and list biosystem components. It is also possible to highlight selected components in a full size image on the source database's web server and to save component lists.
 
BSID: This BSID is the accession number or unique identifier of the biosystem record; it is always an alphanumeric combination (e.g., BSID82291, for human arachidonic acid metabolism).
Related information: The "Related information" menu that appears in the right hand margin of a biosystem record provides a complete set of links to all available data types that are related to this record. Note that some of those data (e.g., genes, proteins, small molecules) are also presented in folder tabs at the bottom of the biosystems record. The folder tabs may present the data differently than the displays produced by the "Related information" menu and/or may provide additional functions not available through the links menus.
|
|
|
|
|
 
Title: The brief title (or name) of the biosystem, for example, "arachidonic acid metabolism", or "citrate cycle", or "TCA cycle", etc. as assigned by the source database.
Type: Record type (organism specific biosystem or conserved biosystem).
Description: The text description of a biosystem provided by the source database. Some biosystem records have a description (e.g., bsid82927, human TCA cycle) and others do not (e.g., bsid82991, human arachidonic acid metabolism).
Organism: The source organism in which a biosystem is found. The organism name, whose Taxonomy ID (TaxID) was provided in the source record, links to the corresponding record in the NCBI Taxonomy database. For additional information, see the data processing/create direct links/taxonomy section of this document.
Source database and corresponding source accession number: The name of the source database that deposited the record into the NCBI BioSystems database, and the local accession number assigned to that record by the source database. For example, the NCBI BioSystem record bsid82991, for human arachidonic acid metabolism, is derived from KEGG record hsa00590 (see illustration).
 
| Thumbnail Image Provided by Source Database |
If a thumbnail image of a biosystem diagram has been provided by a source database, the image is placed in the corresponding BioSystems database record and links to a full-size image that resides on the source database web site. At present, thumbnail images as well as the ability to highlight selected components (genes, proteins, small molecules) in a full size image of the biosystem diagram are currently available only for KEGG records. |
|
| Thumbnail Image Not Available |
If a thumbnail image of a biosystem diagram has not been provided by a source database, a full size diagram may be available at the source database by following the link in the BioSystem record for the source's accession number. |
|
| |
|
| Independent of the availability of a thumbnail image, each BioSystem record lists and categorizes the biosystem components, as noted below, in order to facilitate computation on biosystems data, and provides links to related data throughout the Entrez system. |
|
 
| overview & tips | folder tab descriptions | genes | proteins | small molecules | related biosystems | citations | comments |
Overview & Tips
The folder tabs (annotated illustration) near the bottom of a biosystems record categorize and list biosystem components and associated data. A pagination function at the bottom of each folder tab allows you to browse through a list of up to 3000 items, which include biosystem components specified by the source database as well as genes, protein sequences, and small molecules associated with the biosystem during NCBI data processing).
You can select items on multiple pages within a folder tab (for example, item 3 on page 1 and item 12 on page 2). The selections will persist as you browse within and across folder tabs until you clear the selections in a given tab. However, the Highlight selected records function will work for one folder tab at a time -- the active folder tab.
The "View or save all or selected records in Entrez [DatabaseName]" option allows you to view all (default) or selected records in the database where they reside, and to see the complete list of items if their number exceeds the 3000 item limit of an individual folder tab.
Additional notes are provided below on how to save biosystem component lists and save an archival copy of a biosystem record.
The data displayed by the folder tabs are also accessible via the "Related information" menu in the right hand margin of a biosystem record display (with the exceptions noted below, e.g., Genes and Citations folder tabs). However, some of the folder tabs present the data in a different way (e.g., sort identical proteins by PIG ID in the Proteins folder tab) and/or provide additional functions (e.g., highlight selected components in the source database's full size diagram). Conversely, the "Related information" menu provides access to some data types (e.g., 3D protein structures, biological activity data for small molecules) not presented in the folder tabs; those menus are explained in detail in the "Find related data" section of the help document.
| Folder Tab |
Description and Comments |
| Genes |
 Genes that have been associated with a biosystem using the method descibed in the data processing/create direct links/genes section of this document. Clicking on a GeneID will open the corresponding record in the Entrez Gene database.
Genes that have been associated with a biosystem using the method descibed in the data processing/create direct links/genes section of this document. Clicking on a GeneID will open the corresponding record in the Entrez Gene database.
The check boxes can be used to select multiple records for viewing in the Entez Gene database, or to highlight the selected genes in the full size biosystem diagram at source database. (The "highlight selected records" option will appear in the genes folder tab only if that function is available at the source database.)
If the source record did not specify a GeneID, only the gene name that was specified by the source database in the original record will be displayed and no link to the Entrez Gene database is available. Genes that are shown only by name are visible in the Genes folder tab but are not retrieved by the "Related information : Sequences : Genes" link in the right hand margin of a biosystem record. |
| Proteins |
 Proteins that have been associated with a biosystem using the method descibed in the data processing/create direct links/proteins section of this document.
Proteins that have been associated with a biosystem using the method descibed in the data processing/create direct links/proteins section of this document.
Note that several sequence records might exist for the same protein: primary sequence records submitted to a sequence database by research labs, as well as derivative, specially annotated versions of the sequence created by one or more curated databases such as RefSeq or Swiss-Prot/UniProt. In order to make biosystem records accessible from any version of a protein sequence record, and because the cumulative information in all versions of the sequence record can be greater than the information in any one of the records alone, links are established to all of them. However, to handle this redundancy, protein sequences that are identical in length and composition are clustered at NCBI into protein identity groups (PIGs), and each group is given a PIG ID.
The three display options available in the protein folder tab can be used to show all protein sequence records that have been associated with a biosystem during NCBI data processing, or selected subsets of the protein sequences:
- Non-Redundant Set (default) - lists one representative per protein identity group (PIG). If the PIG includes a RefSeq record, that is selected as the representative. If no RefSeq record is present, then a representative is selected from one the following databases: Swiss-Prot, PIR, PDB, GenPept (protein translations of nucleotide sequence records in GenBank that have been annotated with a coding sequence, or CDS, feature), or PRF, respectively.
- Swiss-Prot/UniProt Only - only Swiss-Prot/UniProt records are listed. If a protein identity group (PIG) contains more than one Swiss-Prot/UniProt, all such members of the PIG will be displayed. If a PIG contains no Swiss-Prot or UniProt record, that PIG will not appear in this view.
- No Filter - lists all protein sequence records from the Entrez Protein database that have been associated with the biosystem during NCBI data processing. Proteins that belong to the same protein identity group (PIG) are grouped together as they are sorted by PIG ID.
The "View or save all or selected records in Entrez Protein" option will retrieve all (default) or selected proteins from the Entrez Protein Database, regardless of which display option is currently active.
If you prefer to only see the list of proteins supplied by the source database for this biosystem, you can save an archival copy of the biosystem record.
Additional note: The check boxes in the folder tab can be used to select multiple records for viewing in the Entez Protein database, or to highlight the selected proteins in the full size biosystem diagram at source database. (The "highlight selected records" option will appear in the proteins folder tab only if that function is available at the source database.)
|
| Small Molecules |
 PubChem Compounds and PubChem Substances that have been associated with a biosystem using the method descibed in the data processing/create direct links/small molecules section of this document.
PubChem Compounds and PubChem Substances that have been associated with a biosystem using the method descibed in the data processing/create direct links/small molecules section of this document. |
| Related BioSystems |
 Other biosystem records that are related to the record you are viewing:
Other biosystem records that are related to the record you are viewing: |
Organism Specific BioSystems |
The organism specific biosystems that correspond, as determined by the source database, to the conserved biosystem currently being viewed. |
Conserved BioSystems |
The conserved biosystem, if one is available, that corresponds to the organism specific biosystem currently being viewed. Conserved biosystems are canonical biosystems that are used to group together orthologous, organism specific biosystems. They are derived from "reference pathways" in the KEGG database and from MetaCyc records. |
Linked BioSystems |
Biosystems that the source database has explicitly stated are connected in some way. |
Similar BioSystems |
Biosystems share at least one identical protein sequence from the same source organism. In other words, the shared protein is in the same protein identity group (PIG) and has the same Taxonomy ID (TaxID). |
Superset BioSystems |
Biosystems that are one node above the one currently being viewed, according to a hierarchical organization used by the source database.
For example, "Phase 1 - Functionalization of compounds" (bsid105699) is a superset of "COX reactions" (bsid105711), reflecting the hierarchical organization of those biosystems specified by the source database, Reactome. |
Subset BioSystems |
Biosystems that are one node below the one currently being viewed, according to a hierarchical organization used by the source database.
For example, "COX reactions" (bsid105711) are a subset of "Phase 1 - Functionalization of compounds" (bsid105699), reflecting the hierarchical organization of those biosystems specified by the source database, Reactome. |
| Citations |
 PubMed records whose PMIDs were explicitly cited in (and extracted from) the source database record. If the source record contains a text citation but no PMID, the citation is displayed in the folder tab, but no link to PubMed is provided and those articles are not retrieved by the the "Related information : Literature : PubMed" link in the right hand margin of a biosystem record.
PubMed records whose PMIDs were explicitly cited in (and extracted from) the source database record. If the source record contains a text citation but no PMID, the citation is displayed in the folder tab, but no link to PubMed is provided and those articles are not retrieved by the the "Related information : Literature : PubMed" link in the right hand margin of a biosystem record. |
| Comments |
 Comments and additional information provided by the source database on the biosystem.
Comments and additional information provided by the source database on the biosystem. |
 
If a source database provides a function to highlight selected biosystem components in the full size diagram on their web site (as KEGG currently does for genes and small molecules), the folder tab for that component type will display an option to "Highlight Selected Records in Source Database." That allows you to see where in a biosystem your gene(s), protein(s), or small molecule(s) of interest occur.
There are two ways to do this: (a) by using check boxes in folder tabs or (b) by writing a specially formatted URL:
(a) Check boxes in folder tabs:
To select specific genes, proteins, or small molecules while viewing a biosystem record, simply activate the check boxes for the component(s) of interest. The pagination function at the bottom of a folder tab allows you to select items on multiple pages within the tab. Then use the "Highlight Selected Records" option to open the full size biosystem diagram on the source database's web site. The components you selected will be displayed with a red outline. Note, however, that the highlight function works for one folder tab at a time -- the active folder tab.
For example, the PTGS1 gene (GeneID 5742) is included in the BioSystems component list for the human arachidonic acid metabolism pathway (bsid82991). If you activate the checkbox for GeneID 5742 in the "Genes" folder tab of the record, then use the option to "Highlight Selected Records in Source Database", the gene will be highlighted with a red outline in the corresponding KEGG record for Arachidonic Acid Metabolism, as shown in the illustration to the right. (In that view, the PTGS1 gene product is displayed as Enzyme Commission (EC) number 1.14.99.1.)
|
|
| HIGHLIGHT SELECTED BIOSYSTEM COMPONENTS |
|
|
(b) Specially formatted URL:
Another way to select components of interest is to write a URL using the following syntax:
http://www.ncbi.nlm.nih.gov/biosystems/bsidnumber?Sel=categoryA:uid1,uid2,uid3;categoryB:uid4,uid5;categoryC:uid6,uid7#show=foldertabname
The bsidnumber should be entered as digits only. Category values can be geneid (Entrez Gene IDs), gi (Protein GI numbers), or cid (PubChem Compound IDs). If two or more categories are included in a URL, they must be separated by a semicolon as shown in example 1 below. Comma separated unique identifiers (uids) should be provided in the "Sel" (select) parameter. The anchor tag (#show=foldertabname) can be used to specify which folder should be displayed as the default tab on the biosystem summary page; only one default folder tab can be specified in a URL and allowable values are #show=genes, #show=proteins, and #show=smallmolecules. If the anchor tag is not present in the URL, the genes folder tab will be displayed by default. In that case, selections you made in other folders can be viewed simply by clicking on their tabs.
Example 1: Select the specified genes, protein, and compound within BioSystem record bsid82991, and display the "Small Molecules" folder tab by default:
http://www.ncbi.nlm.nih.gov/biosystems/82991?Sel=geneid:5742,5743;gi:189774;cid:445049#show=smallmolecules
Example 2, 3a&b, 4a&b: Select item(s) within only one component category. The #show anchor specifies which folder tab to display as the default. If that parameter is omitted from the URL, the Genes folder tab will be displayed by default:
http://www.ncbi.nlm.nih.gov/biosystems/82991?Sel=geneid:5742,5743
http://www.ncbi.nlm.nih.gov/biosystems/82991?Sel=gi:189774 http://www.ncbi.nlm.nih.gov/biosystems/82991?Sel=gi:189774#show=proteins
http://www.ncbi.nlm.nih.gov/biosystems/82991?Sel=cid:445049
http://www.ncbi.nlm.nih.gov/biosystems/82991?Sel=cid:445049#show=smallmolecules
Additional notes: (1) The highlight function is currently only available for KEGG records. If the URL contains a BSID for a record from a different source database, the URL will open the BioSystems record and activate the check boxes of the items you have selected in each component category; however, the folder tabs will not contain the "Highlight Selected Records" option. (2) If a URL contains invalid component UIDs (i.e., gene, protein, or compound IDs that are not present in the specified biosystem), the biosystem record display will select/highlight only the subset of UIDs that are present. UIDs that are absent from the biosystem will also be absent from the corresponding folder tab, and no explicit error message will list the absent UIDs.
|
|
 
Biosystem component lists can be saved by following the "Click to view and/or save record in Entrez [DatabaseName]" link in the folder tab of the desired component type. Doing this will retrieve the genes/proteins/small molecules that were explicitly listed by the source database in their original biosystem record, plus additional genes/proteins/small molecules that have been associated with the biosystem by the NCBI data processing procedure.
Once you are viewing the components in the relevant Entrez database, you can display and/or save those records in any format that is available for that database. For example, records from the Entrez Protein database can be saved in FASTA format (which is convenient for sequence analysis), as a list of GI numbers, or in other formats such as GenPept (which contains sequence data plus annotations, similar to GenBank format). The Entrez help document provides additional information about sequence database record formats. The Entrez Gene help and PubChem help documents describe record formats for genes and small molecules, respectively.
 
| |


 |
The ASN.1, XML, and JSON buttons near the bottom of a biosystem record save an archival copy of the biosystem record that reflects the content deposited by the source database. This includes only the genes/proteins/small molecules that were explicitly listed by the source database in their deposited record, and not the additional genes/proteins/small molecules that were associated with the biosystem record via NCBI data processing. The archival record also includes the additional content that was present in the deposited record, such as annotations about the biosystem. |
|
|
|
|
|
|
|
 |
|
|
|
|
 
 
The articles below are provided for your convenience if you would like more information on any source database. Please consult the web site of a source database for information on how to cite it and for additional articles.
 |
BioCyc & MetaCyc -- Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Paley S, Popescu L, Pujar A, Shearer AG, Zhang P, Karp PD. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010 Jan; 38(Database issue): D473-9. (Epub 2009 Oct 22) [PubMed PMID: 19850718] [Full Text] [Database Websites: http://biocyc.org/ and http://metacyc.org/] |
 |
EcoCyc -- Keseler IM, Bonavides-Mart�nez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, Peralta-Gil M, Santos-Zavaleta A, Shearer AG, Karp PD. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009 Jan; 37(Database issue): D464-70. [PubMed PMID: 18974181] [Full Text] [Database Website: http://ecocyc.org/] |
 |
Gene Ontology (GO) -- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000 May;25(1):25-9. [PubMed PMID: 10802651] [Full Text] [Database Website: http://www.geneontology.org/] |
 |
KEGG -- Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010 Jan; 38(Database issue): D355-60. (Epub 2009 Oct 30) [PubMed PMID: 19880382] [Full Text] [Database Website: http://www.genome.jp/kegg/] |
 |
LIPID MAPS (online tools) -- Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007 Jul; 35(Web Server issue):W606-12. Epub 2007 Jun 21. [PubMed PMID: 17584797] [Full Text] [Database Website: http://www.lipidmaps.org/tools/] |
 |
LIPID MAPS (structure database) -- Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH Jr, Murphy RC, Raetz CR, Russell DW, Subramaniam S. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007 Jan; 35(Database issue): D527-32. (Epub 2006 Nov 10) [PubMed PMID: 17098933] [Full Text] [Database Website: http://www.lipidmaps.org/data/structure/] |
 |
PID -- Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009 Jan; 37(Database issue): D674-9. (Epub 2008 Oct 2) [PubMed PMID: 18832364] [Full Text] [Database Website: http://pid.nci.nih.gov/] |
 |
Reactome -- Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, Kanapin A, Lewis S, Mahajan S, May B, Schmidt E, Vastrik I, Wu G, Birney E, Stein L, D'Eustachio P. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009 Jan; 37(Database issue): D619-22. [PubMed PMID: 18981052] [Full Text] [Database Website: http://www.reactome.org/] |
 |
Reactome -- Vastrik I, D'Eustachio P, Schmidt E, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, Matthews L, Wu G, Birney E, Stein L. Reactome: a knowledge base of biologic pathways and processes Genome Biology 2007 Mar; 8(3):R39. [PubMed PMID: 17367534] [Full Text] [Database Website: http://www.reactome.org/] |
 |
WikiPathways -- Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008 Jul 22;6(7):e184. [PubMed PMID: 18651794] [Full Text] [Database Website: http://www.wikipathways.org/index.php/WikiPathways] |
|
|
|
|







 As noted in the page on
As noted in the page on