BLASTFeed
Improved BLAST statistics described in BMC Research Notes.
Fri, 11 Jan 2013 10:00:00 EST
nt will become default BLAST db
Fri, 16 Nov 2012 14:00:00 EST
Starting November 26, 2012, the nucleotide collection (nt) will be the default nucleotide search database. The nucleotide collection consists of GenBank+EMBL+DDBJ+PDB+RefSeq sequences, but excludes EST, STS, GSS, WGS, TSA, patent sequences as well as phase 0, 1, and 2 HTGS sequences.
The nt database recently joined a list of other fast, indexed database searches offered by the NCBI that include the human G+T (genome plus transcript) and mouse G+T databases, as well as the human and mouse reference genome databases. The indexed databases are available when using megaBLAST. Indexed megaBLAST, at the NCBI BLAST web page, can search queries of a couple thousand bases against the 43 billion base nt database in a few seconds.
Indexed searches at the NCBI use an in-memory index described by Morgulis et al. and the megaBLAST algorithm.
.New BLAST report
Wed, 31 Oct 2012 16:00:00 EST
BLAST 2.2.27+ released
Mon, 10 Sep 2012 14:00:00 EST
1.) We have implemented composition-based statistics for BLASTX. It offers better statistical accuracy and is the default mode, but the older behavior can be recovered with the –comp_based_stats flag. The BLASTX implementation is based upon the methods described in pmid 17156431.
2.) The deltablast application now allows searches with the –remote flag.
3.) We have reduced the memory usage of the blastn application when searching many small queries. This involved reducing the number of queries searched during one pass through the database as well as reducing the number of database sequences retrieved for the tabular output formatting. Section 4.3.1 and 4.3.2 of the manual present details on this change as well as information on how to control the number of queries searched during one pass through the database.
4.) The output for reports without separate descriptions and alignments sections (all –outfmt greater than 4) should now use –max_target_seqs to control the output rather than –num_descriptions and –num_alignments.
5.) In megaBLAST mode, the blastn application now reduces the number of gaps if possible. As described in pmid 10890397, the megaBLAST algorithm (with linear gapping) assigns an equal score to an alignment with two mismatches and to an alignment with two gaps plus an additional match. If possible, the blastn application now presents the alignment without the gaps.
6.) We have extensively rewritten the BLAST+ manual. Appendix C now contains tables of options for different programs. This is an updated set of tables based on the supplementary information of pmid 20003500.
7.) We have fixed various bugs in the blast_formatter, blastdbcmd and other programs. These include one that did not render an asterisk (stop codon) properly as well as one that improperly applied compostion-based statistics to any use of the Smith-Waterman option. .
Improved BLASTX statistics
Wed, 01 Aug 2012 17:00:00 EST
BLASTX searches translate a nucleotide query and compare it to a protein database. BLASTX searches with CBS produce more reliable results. The increased accuracy is especially noticeable for query and subject sequences with biased compositions (i.e., low-complexity sequences). CBS has been available for some time with BLASTP, PSI-BLAST, and TBLASTN.
CBS is enabled by default for BLASTX searches at the NCBI BLAST website. CBS may be disabled by using the "Composition adjustment menu" in the Algorithm parameters part of the page. CBS will be available in the 2.2.27+ release of stand-alone BLASTX.
The CBS implementation for BLASTX is similar to that of TBLASTN, described at http://www.ncbi.nlm.nih.gov/pubmed/17156431.
OLD_BLAST URL to be discontinued. Alternative NCBI BLAST parsable formats are available
Thu, 12 Jul 2012 16:00:00 EST
For information on parsable formats, please see http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=DeveloperInfo and the links on that page.
The standard BLAST report is a human readable format that is subject to change with little or no notice. In order to facilitate coming enhancements to the BLAST report, it is necessary to discontinue support for the OLD_BLAST URL parameter. The OLD_BLAST parameter is currently used by a very small fraction of interactive users and results in an older version of the BLAST report (in HTML). The BLAST web pages will simply ignore the OLD_BLAST parameter starting September 10, 2012.
Please address any concerns or questions to blast-help@ncbi.nlm.nih.gov..
Primer-BLAST article in BMC Bioinformatics
Thu, 05 Jul 2012 16:00:00 EST
The article is available through http://www.ncbi.nlm.nih.gov/pubmed/22708584 and discusses the design and implementation of Primer-BLAST.
The Primer-BLAST web page is at http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi.
Microbial BLAST
Mon, 04 Jun 2012 12:00:00 EST
The microbial BLAST page has been redesigned for ease of use and better integration with other BLAST pages. The page now allows selection of taxonomic catgegories through an auto-complete mechanism (start typing in "Organism" box and select a suggestion). Multiple taxonomic categories can be included or excluded. For nucleotide databases, the search sets have also been divided into Complete and Draft genomes. Other standard features of the BLAST pages such as "Edit and Resubmit" (start a new search from a BLAST report with current settings) and the ability to optimize for a specific search (under "Program Selection") are available.
The old microbial page will be available at http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi at least until the end of June, 2012..
DELTA-BLAST
Mon, 23 Apr 2012 12:00:00 EST
Since 1997, the BLAST web site has offered searches with a “Position Specific Scoring Matrix” (PSSM) through the PSI-BLAST program. This normally required the user to launch a few searches. Domain Enhanced Lookup Time Accelerated BLAST (DELTA-BLAST) is a new addition to the web site that performs a PSSM search. It runs a fast RPSBLAST search in order to construct the PSSM and then searches the PSSM against a BLAST database. Tests using the SCOP based benchmark set demonstrate that DELTA-BLAST yields retrieval accuracy greater than BLASTP and similar to a few PSI-BLAST iterations. The DELTA-BLAST results can also be used to initiate a PSI-BLAST search.
You may start a DELTA-BLAST search from the protein-protein page.
An article in Biology Direct describes the DELTA-BLAST algorithm and benchmark results..
BLAST 2.2.26+ release.
Thu, 01 Mar 2012 14:00:00 EST
BLAST+ applications may be downloaded here. Instructions on the installation of BLAST+ applications are available here.
The BLAST+ 2.2.26 release contains a number of important changes and improvements:
1.) DELTA-BLAST. A new application called deltablast is included in this release. Deltablast stands for Domain Enhanced Look-up Time Accelerated BLAST. It first uses RPS-BLAST to align a protein query to conserved domains in CDD, then performs a sequence database search using a position specific score matrix (PSSM) derived from the aligned domains. The PSSM construction method is similar to that of PSI-BLAST, but begins by aligning the query to CD's rather than to individual sequences. DELTA-BLAST can be much more sensitive than standard BLASTP. DELTA-BLAST is also available from the "protein blast" link at blast.ncbi.nlm.nih.gov. DELTA-BLAST needs a special version of CDD database that contains some extra files. Instructions for downloading and installing this specialized copy of the CDD database can be found in section 5.18 of the BLAST Command Line Application Manual at http://www.ncbi.nlm.nih.gov/books/NBK1763/
2.) New finite size correction (FSC). The FSC is subtracted from the query and database sequence length for the calculation of the BLAST statistics used to rank the results. The older FSC did not properly handle short query or database sequences, as the estimated FSC might be longer than a short sequence, and it was necessary to simply set the resulting length to an ad hoc value (typically one). The new approach elides this issue by looking at the expected values of both the query and database length together, rather than separately. In general, it ranks matches involving a short query or database sequence as more significant. The new FSC increases the ROC score (at 4853 FP) found with a SCOP test set by about 2%. For short queries or database sequences, it may change the expect value reported by orders of magnitude. Currently, the new FSC is only implemented for protein-protein programs (e.g., blastp, psiblast, blastx, rpsblast, etc.), but not the blastn application. The old behavior may be recovered by setting the environment variable OLD_FSC to a non NULL value.
3.) Makeprofiledb. Makeprofiledb can be used to make search sets for RPS-BLAST, including the specialized data needed by DELTA-BLAST. Makeprofiledb is a replacement for the C toolkit application formatrspdb.
4.) Blastcl3 users should switch to BLAST+. Blastcl3 is deprecated and the service will need to be retired in the not too distant future. This client and service have served the community well since 1997, but changes in the way BLAST searches are done at the NCBI (e.g., a Request ID can be issued for a search) mean that a better and more robust client can be offered. The BLAST+ applications can send off remote searches if the argument -remote is added. More details are available at http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastNews
5.) Last C toolkit binary release. This is the last release of the C toolkit BLAST binaries (e.g., blastall, blastpgp, etc.). The source code for these application is not being updated anymore, but will continue to be available. Users of these legacy binaries are encouraged to move to the BLAST+ applications that are being actively developed. Help on transitioning to the BLAST+ applications can be found at http://www.ncbi.nlm.nih.gov/books/NBK1763/
ChangeLog:
-----------------------------
* 2.2.26 release.
* Mac executables are now Universal Binaries for 32- and 64-bit architectures, we no longer produce PPC and Intel Universal binaries. The executable archive names remain unchanged.
* Added DELTA-BLAST - a new tool for sensitive protein searches
* Added makeprofiledb - a tool for creating a database for RPS-BLAST
Improvements:
* The blast_formatter application can now format bl2seq RIDs.
* PSI-BLAST can produce archive format, blast_formatter can format that output.
* PSI-BLAST has two new options that work with multiple-sequence alignments: ignore_msa_master and msa_master_idx (see BLAST+ manual).
* mkmbindex can now create masked indices from a BLAST database and ASN.1 masking data.
* An improved finite size correction is now used for blastp/blastx/tblastn/rpsblast. The FSC is subtracted from the query and database sequence length for the calculation of the expect value. The new FSC results in more accurate expect values, especially for alignments with a short query or target sequence. Re-enable the old size correction by setting the environment variable OLD_FSC to a non-NULL value.
* The blastdbcmd -range parameter now accepts a blank value for the second parameter to signify the end of a sequence (e.g., -range "100-")
* There was a performance improvement for long database sequences in results with many matches.
Bug fixes:
* There was a blastn problem if subject_loc and lcase_masking were used together.
* There was a problem with multi-threaded blastx if the query included a long (10,000+) sequence of N's.
* The percent identity calculation was wrong if the best-hit algorithm was used.
* There was a problem with the multiple BLAST database statistics report in XML format.
* Makeblastdb failed to return an error when input was not available.
* The formatting option -outfmt "7 nident" always printed zero.
* The search strategy was not properly saving the -db_soft_mask option.
* An error message was emitted if there was a "<" in the query title.
* A problem reading lower-case masking from the query could cause a search to fail.
.
BLAST+ remote searches.
Fri, 24 Feb 2012 09:00:00 EST
The BLAST+ applications can perform remote searches.
The BLAST+ applications can not only run searches on local machines, but also perform searches on the NCBI servers. Sending searches to the NCBI servers can be advantageous if you have not downloaded the BLAST databases or do not have sufficient resources for a set of searches. Please run only one remote instance of the BLAST+ application at a time.
Information on downloading, installing and using the BLAST+ applications is available at http://www.ncbi.nlm.nih.gov/books/NBK1762/ To enable remote searches, simply add -remote to any BLAST+ command-line.
The BLAST+ remote service replaces the older blastcl3 application. This new service has a number of advantages over the blastcl3 application. Blastcl3 requires a persistent connection during the entire search, can only submit one query at a time, and is unable to return the BLAST Request ID (RID) used in the search. The BLAST+ remote service can submit multiple queries (from FASTA input) at once, poll for the results using the BLAST RID, and also print the RID in the BLAST report. Using the BLAST RID, it is possible to reformat the search with the blast_formatter application, reformat the search at the NCBI web site, or use analysis tools such as the BLAST treeview or the taxonomy report.
On March 5, 2012, the blastcl3 client will start printing a reminder to standard error that the service is deprecated and that users should change to the BLAST+ applications. The reminder will not interfere with the search or the formatting of results.
.BLAST database list
Wed, 01 Feb 2012 09:00:00 EST
1.) The WGS database on the nucleotide blast, tblastn and tblastx pages now allows taxonomic limits with an auto-complete menu (start typing and look at the suggestions).
2.) The est_others database has been removed. Instead, use the taxonomic auto-complete menu to search a subset of the est database.
3.) The env_nt database has been removed. These sequences are actually WGS metagenomic sequences. They can now be searched by selecting the WGS database and limiting the search (with the taxonomic auto-complete menu) to "metagenome".
4.) A new 16S database is now available through the main nucleotide BLAST page. See the NCBI News for details.
5.) The env_nr database has been renamed "Metagenomic proteins".
Note that the new WGS functionality makes the WGS link under specialized BLAST redundant and it has been removed. .
SOAP BLAST
Mon, 18 Jul 2011 08:00:00 EST
Genomic BLAST page update
Thu, 19 May 2011 11:00:00 EST
Transcriptome Shotgun Assembly database
Tue, 10 May 2011 13:00:00 EST
BLAST 2.2.25 release
Thu, 24 Mar 2011 09:00:00 EST
Users are encouraged to use the BLAST+ applications available at ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/ This release includes a substantial number of bug fixes and new features for the BLAST+ applications.
Improvements:
* Enhanced documentation, includes simplified setup instructions, available at http://www.ncbi.nlm.nih.gov/books/NBK1762
* Added support for hard-masking of BLAST databases.
* Improve performance of makeblastdb for FASTA input with large numbers of sequences, improve error checking.
* Allow Best Hit options and XML formatting for Blast2Sequences mode
* Allow multiple query sequences for psiblast.
* Allow specification of any multiple sequence alignment sequence as the master with the -in_msa psiblast argument.
* Add an optional -input_type argument to makeblastdb.
* Added support for query and subject length to tabular output.
* Performance of -seqidlist argument improved.
* The minimum of the number of descriptions and alignments is now used for tabular and XML output (consistent with the behavior of the older blastall applications).
Bug fixes:
* Makeblastdb and blastdbcmd problems with parsing, storing, and retrieving sequence identifiers.
* Missing subject identifiers in tabular output.
* Blast_formatter ignoring -num_alignments and -num_descriptions
* Blast archive format could be saved incorrectly with multiple queries.
* Blast_formatter established an unneeded network connection.
* Blast_formatter did not save masking information correctly.
* Rpstblastn might crash if searching many sequences.
* Indexed megablast would not run in multi-threaded mode.
* Query title in the PSSM saved by psiblast was not being stored.
* Possible failure to run in multi-threaded mode with multiple queries or large database sequences.
* Tblastn runs with database masking might miss matches.
* Truncated output for sequence input with extra spaces in the defline
* Problem with MacOSX binaries on MacOSX 10.5
BLAST+ applications, as well as the legacy C applications (e.g. blastall), may be downloaded from http://www.ncbi.nlm.nih.gov/blast/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download .
New SNP BLAST page
Wed, 12 Jan 2011 14:00:00 EST
The SNP blast databases are built from RefSNP(rs) flanking sequences as their sequence source. The RefSNP set includes not only polymorphisms but also rare clinical variants from human. In addition to single nucleotide variants, SNP blast databases also include flanks from insertions/deletions, short tandem repeats and multi-base nucleotide variations. Typical use cases for dbSNP BLAST page includes:
1.) Confirming that a novel SNP has not been reported in dbSNP using your flanking sequences.
2.) Determining if a sequence you have overlaps flanks from or includes variations stored in dbSNP.
.New WGS BLAST page
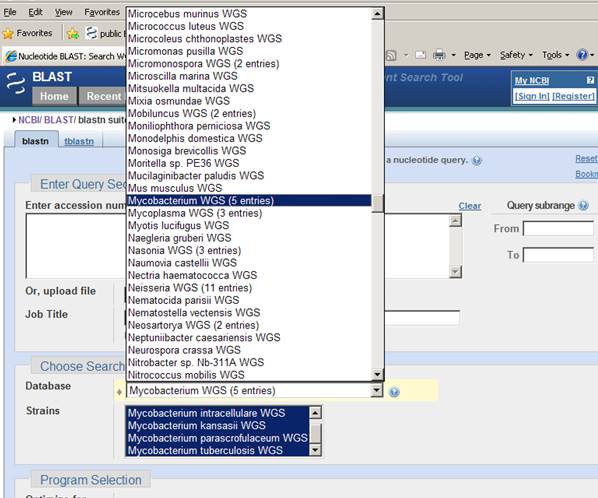
Mon, 22 Nov 2010 09:00:00 EST
The main menu on the new page lists search sets by genus, or by species if only one species in a genus is available. A sub-menu allows selection of one or more of the species from a given genus. The example below shows the genus Mycobaterium selected/ There are five species available for this genus.
makeblastdb update available
Mon, 13 Sep 2010 11:00:00 EST
The makeblastdb application distributed with the BLAST+ 2.2.24 release has a very significant performance problem when the -parse_seqids option is used. Patched binaries for common platforms are available at ftp://ftp.ncbi.nlm.nih.gov/blast/executables/snapshot/2010-09-13/.
BLAST 2.2.24 release
Mon, 23 Aug 2010 13:00:00 EST
This release includes a number of bug fixes as well as new features for the BLAST+ applications:
* Introduced the BLAST Archive format to permit reformatting of stand-alone BLAST searches with the blast_formatter (see BLAST+ user manual)
* Added the blast_formatter application (see BLAST+ user manual)
* Added support for translated subject soft masking in the BLAST databases
* Added support for the BLAST Trace-back operations (btop) output format
* Added command line options to blastdbcmd for listing available BLAST databases
* Improved performance of formatting of remote BLAST searches
* Used a consistent exit code for out of memory conditions
* Fixed bug in indexed megablast with multiple space-separated BLAST databases
* Fixed bugs in legacy_blast.pl, blastdbcmd, rpsblast, and makeblastdb
* Fixed Windows installer for 64-bit installations
BLAST+ applications, as well as the legacy C applications (e.g. blastall), may be downloaded from http://www.ncbi.nlm.nih.gov/blast/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download.
BLAST 2.2.23 release
Mon, 22 Mar 2010 15:00:00 EST
This release includes a number of bug fixes for the BLAST+ applications:
1.) BLASTN runs with word-size four were missing hits in the 2.2.22 release, this has been fixed.
2.) Query/subject offsets for tabular formatting (query on minus strand) was fixed.
3.) A MEGABLAST performance regression (with query masking) was fixed.
4.) Seg filtering for long blastx queries was failing. This has been fixed and optimized.
5.) A bug that caused tabular output to contain tokens like "gnl|BL_ORD_ID|1 " was fixed.
6.) Search strategies can now be used in bl2seq mode.
7.) Problems with displaying accessions in XML output have been fixed.
8.) Problems with percent identity and percent positive matches in the the tabular output have been fixed.
BLAST+ applications, as well as the legacy C applications (e.g. blastall), may be downloaded from http://www.ncbi.nlm.nih.gov/blast/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download .
COBALT improvements
Thu, 21 Jan 2010 17:00:00 EST
The following popular formats, used by alignment viewers as well as sequence and phylogeny analysis tools, are supported: FASTA plus gaps, ClustalW, Phylip, and Nexus. The alignment can also be downloaded as a Seq-align in ASN.1 format.
In order to download your alignment click the 'Download' link on the top of the results page and then select desired format in the 'Download alignment' box.
COBALT is a multiple alignment program for proteins that can be accessed from http://www.ncbi.nlm.nih.gov/tools/cobalt/cobalt.cgi or BLASTP search results at the NCBI.
The COBALT algorithm is described in this paper.
.BLAST+ article in BMC Bioinformatics
Fri, 18 Dec 2009 08:00:00 EST
The BMC Bioinformatics article is available from http://www.biomedcentral.com/1471-2105/10/421/abstract and discusses changes to search chromsome length database sequences and long queries faster. The article also describes database masking, a more modular design, and new command-line applications.
The new command-line applications are available at http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download.
Exclude Models and environmental samples
Fri, 06 Nov 2009 10:00:00 EST
BLAST 2.2.22 now available
Mon, 19 Oct 2009 11:00:00 EST
This release includes new BLAST+ command-line applications.
The BLAST+ applications have a number of advantages over the older applications and users are encouraged to migrate to the new applications. The new applications can be downloaded from ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST These applications have been built with the NCBI C++ toolkit. Changes from the last release are listed below.
The older C toolkit applications (e.g., blastall) are still available at ftp://ftp.ncbi.nlm.nih.gov/blast/executables/release/2.2.22/
Changes from the last release are listed below.
Please send questions or comments to blast-help@ncbi.nlm.nih.gov
Changes for the BLAST+ applications:
* Added entrez_query command line option for restricting remote BLAST databases.
* Added support for psi-tblastn to the tblastn command line application via
the -in_pssm option.
* Improved documentation for subject masking feature in user manual.
* User interface improvements to windowmasker.
* Made the specification of BLAST databases to resolve GIs/accessions
configurable.
* update_blastdb.pl downloads and checks BLAST database MD5 checksum files.
* Allow long words with blastp.
* Added support for overriding megablast index when importing search strategy
files.
* Added support for best-hit algorithm parameters in strategy files.
* Bug fixes in blastx and tblastn with genomic sequences, subject masking,
blastdbcheck, and the SEG filtering algorithm.
Changes for C applications:
* Blastall was not able to use BLAST databases with only accessions to format results, this has been fixed.
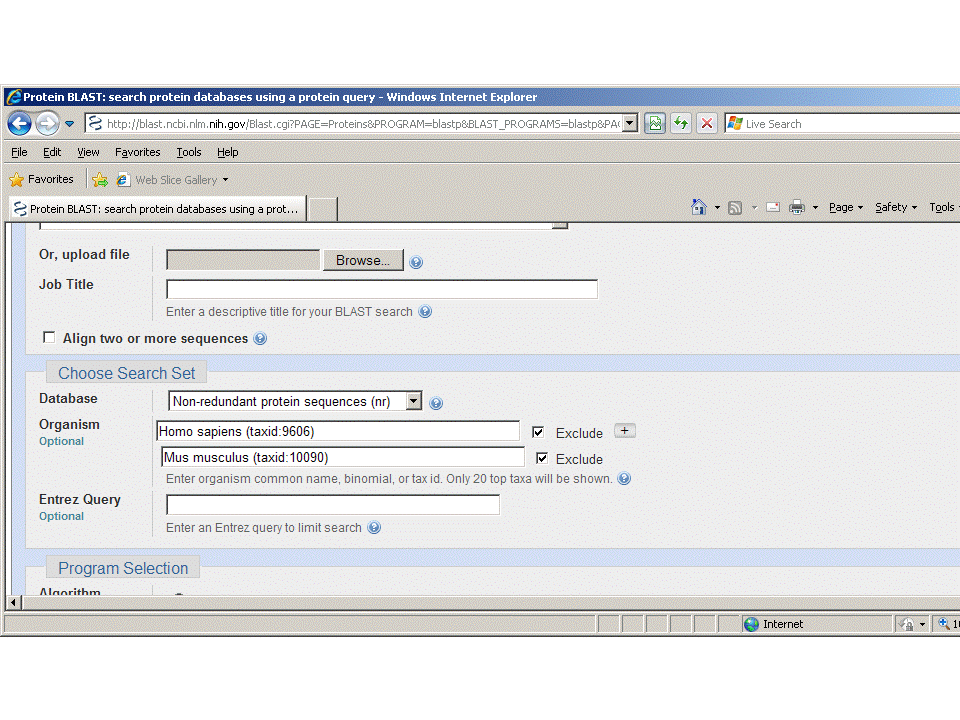
.Limit by organism improved
Mon, 14 Sep 2009 09:00:00 EST
 .
.
BLAST 2.2.21 now available
Tue, 28 Jul 2009 11:00:00 EST
This release includes new BLAST+ command-line applications.
The BLAST+ applications have a number of advantages over the older applications that include working more robustly with long sequences and a new type of masking (database masking). For details see ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/user_manual.pdf. The new applications can be downloaded from ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST These applications have been built with the NCBI C++ toolkit. Changes from the last release are listed below.
The older C toolkit applications (e.g., blastall) are still available at ftp://ftp.ncbi.nlm.nih.gov/blast/executables/release/2.2.21/
Changes from the last release are listed below.
Please send questions or comments to blast-help@ncbi.nlm.nih.gov
c toolkit binary changes:
* corrected a bug in xml output (SB-217)
* corrected a bug with query concatenation in ungapped searches (SB-263)
* tabular output header for "-m 8" now printed even if there are no results. (sb-290)
C++ toolkit binary improvements:
* best hit algorithm, see section 4.5.12 in ftp://ftp.ncbi.nih.gov/blast/executables/blast+/LATEST/user_manual.pdf
* improve culling option performance
* fix mutex problems in BLAST database reader.
* improve performance of database masking option.
C++ binary changes:
* database masking enabled, see details in ftp://ftp.ncbi.nih.gov/blast/executables/blast+/LATEST/user_manual.pdf
* makeblastdb user-interface improvements
* blastdbcmd can now emit masked fasta for a masked database
.Multiple Alignments with COBALT
Mon, 06 Jul 2009 13:30:00 EST
(http://www.ncbi.nlm.nih.gov/pubmed/17332019).
COBALT can be started from BLAST results (use the "Multiple Alignment" link) or from the COBALT web site at http://www.ncbi.nlm.nih.gov/tools/cobalt/cobalt.cgi?CMD=Web .
SRA transcript BLAST
Mon, 27 Apr 2009 11:00:00 EST
The search sets are grouped by organism and include all public 454 transcript sequences in the NCBI SRA database. Go to http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=MegaBlast&PROGRAM=blastn&BLAST_PROGRAMS=megaBlast&PAGE_TYPE=BlastSearch&BLAST_SPEC=SRA to run a search.
BLAST 2.2.20 now available
Fri, 03 Apr 2009 16:00:00 EST
1.) Ungapped blastn searches allow arbitrary reward/penalty scores.
2.) Spaces are allowed in database pathnames on windows
3.) Seedtop now has gilist support.
4.) Fix a bug that caused the number and order of queries to affect blastx results.
5.) Modified the 2-hit blastn algorithm so that no overlap is allowed between hits.
Align two sequences form.
Tue, 03 Feb 2009 16:00:00 EST
BLAST 2.2.19 now available
Wed, 17 Dec 2008 11:00:00 EST
- The BLASTDB environment variable now supports multiple database search paths.
- When possible, a smaller protein lookup table is used to improve performance.
- formatrpsdb now supports creating databases larger than 2G.
- seedtop now supports searches with gi lists.
- The X3 value for blastn/megablast was corrected.
Align Sequences with BLAST
Thu, 04 Sep 2008 11:00:00 EST
Find specific primers with Primer-BLAST
Tue, 22 Jul 2008 11:00:00 EST
To get started with Primer-BLAST go to http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastNews and enter FASTA, an accession or a GI into the PCR Template box. Or alternately fill in the forward and reverse primers in the "Primer Parameters" area. By default human sequences are searched in the specificity check, but that may be changed to other organisms in the " Primer Pair Specificity Checking Parameters". Use the "Get Primers" button at the bottom of the page to submit your search.
Please send question and comments about Primer-BLAST to blast-help@ncbi.nlm.nih.gov
.
BLAST interface described in NAR web server issue
Fri, 27 Jun 2008 11:00:00 EST
New tree view available
Thu, 12 Jun 2008 11:00:00 EST
- The tree can be downloaded in Newick or Nexus format recognized by popular phylogenetic software packages.
- The tree can be rooted at any user-selected node. This option is available from the node pop-up menu.
- Any user-selected subtree can be collapsed into a single node.
- Sub-trees with sequences from only one Blast Name are automatically collapsed (a Blast Name is a high level taxonomic grouping). This behavior can be controlled by the "Collapse Mode" menu on the right side of the tree view page..
BLAST report improvements
Mon, 12 May 2008 11:00:00 EST
New BLAST URL available
Fri, 25 Apr 2008 11:00:00 EST
New BLAST Redesign in Production
Fri, 13 Apr 2007 14:00:00 EST
----------------------------------------------------------------
The new NCBI BLAST pages will become the default interface at
http://ncbi.nlm.nih.gov/blast on April 16, 2007. The new
interface is currently available as a beta release at
http://ncbi.nlm.nih.gov/blast/beta/. For details on the new
interface, see http://www.ncbi.nlm.nih.gov/BLAST/beta/about/.
After the new interface is released, the previous interface will
remain available from a link on the new front page until May 14,
2007.
A Note About URLAPI
The new BLAST pages support URLAPI, a protocol that scripts and
programs use to run BLAST searches and retrieve results over
HTTP. (For more on URLAPI, see
http://www.ncbi.nlm.nih.gov/blast/Doc/urlapi.html). The following
information only applies to you if you develop or are responsible
for software that uses URLAPI.
The new pages have been tested and produce correct results with
the following URLAPI client programs:
* the BioPERL RemoteBlast module
* the NCBI demo script http://ncbi.nlm.nih.gov/blast/docs/web_blast.pl
* various scripts used in-house at NCBI
Users of URLAPI should be aware of the following minor
changes. In the new interface:
1. The Request ID (RID) format will be shorter. The new format
is 11 alphanumeric characters (e.g. RDEFEA5012) and will have no
internal structure. The previous RID format was 36 or more
characters long, including punctuation (e.g.,
1175172712-21345-42512597310.BLASTQ3).
2. BLAST reports will show masked regions as lower-case letters
by default (see
http://nar.oxfordjournals.org/cgi/content/full/34/suppl_2/W6,
figure 2. The current default behavior is to show masked
regions as N's or X's. Users may recover the current behavior
by adding &MASK_CHAR=0 to the query string for a URLAPI
request.
3. BLAST reports will show alignments for 100 database sequences
by default. The current reports show only 50 alignments by
default.
If you have any questions please send them to mcginnis at ncbi.nlm.nih.gov.
Special Announcement: Beta Test of New BLAST Interface
Mon, 05 Mar 2007 14:00:00 EST
NCBI is holding a beta test for a new BLAST interface design. We invite you to try these pages and send us your comments and suggestions.
One major improvement is a new "Recent Results" feature that provides links to all of your recent BLAST search results. Another is "Saved Strategies", which allows you to save BLAST forms with their parameters and use them later. Saved Strategies requires a free MyNCBI account, and is compatible with existing accounts. Signing in to MyNCBI also makes your Recent Results available from any browser.
Other improvements include:
- Easier navigation
- Simplified BLAST program selection
- Easy access to genome searches
- Improved Organism selection with species name auto-complete
- Automatic parameter adjustment to optimize for short queries
- A user-specifiable title for each BLAST job
The Beta test is available at:
http://www.ncbi.nlm.nih.gov/blast/beta/
or as a link from the BLAST home page at
http://www.ncbi.nlm.nih.gov/blast/
Please send all comments suggestions or bug reports to
mcginnis@ncbi.nlm.nih.gov.
BLAST 2.2.14 now available
Wed, 07 Jun 2006 00:05:00 EST
Major Changes
blastall now uses the new engine by default, resulting in significant performance improvements\ and enabling query concatenation for all program types.BLAST 2.2.13 now available
Sun, 12 Jun 2005 12:00:00 EST
Major changes
New engine available in blastall
Blastall now has support for a new version of the BLAST engine that can be enabled by adding"-V F" to the blastall command-line. This option will probably be the default in future versions. There are a few situations where it is very advantageous to use the new engine:Statistical parameter change
Megablast, blastall and bl2seq have until now allowed users to select arbitrary gap existence and extension penalties for a blastn type search. This has been convenient for users but has led to the unfortunate situation that searches with some parameter sets were significantly overestimating the statistical significance of matches. To address this problem the proper statistical parameters for a number of reward/penalty/gap existence/gap extension values have been calculated. The parameters that might cause an issue here are-r (match reward), -q (mismatch penalty), -G (gap existence cost), and -E (gap extension cost). If you do not change these, then nothing will change for you. Please email blast-help@ncbi.nlm.nih.gov with any questions, bug reports, or requests for different parameter sets.