A Closer Look
Singled Out: Researchers Look to Single Cells for Cancer Insights
When asked about the biggest challenges to better understanding cancer, one word practically leaps from the mouths of many researchers: heterogeneity.
A tumor, the researchers stress, is not a uniform mass of identical cells with identical behaviors. Cells can act quite differently in one part of a tumor than in another. Genes critical to cell proliferation, for instance, may be active in one area but not another, or a subpopulation of cancer cells may be dormant, practically hiding from any drug that may try to enter their lair.
This heterogeneity has been blamed, for example, for the limited success of targeted therapies and of efforts to identify better diagnostic and prognostic markers of disease.
Researchers are now discovering what many have long suspected: much of what makes tumors heterogeneous overall is the substantial heterogeneity among individual cancer cells.
Until recently, the meticulous scrutiny of individual cells has been nearly impossible, particularly because of the relative scarcity in each cell of the key components that need to be measured, such as DNA and RNA. But thanks to technological advances that can help overcome some of those limitations, a growing number of investigators are beginning to delve deeper into the biology of the single cell.
The studies conducted to date “show us how much diversity there is among cancer cells in a given tumor,” said Dr. Garry Nolan, an immunologist at Stanford University whose lab is focused on mapping communication networks in individual cancer cells.
Even with improved technology, however, conducting studies at the single-cell level is difficult and can be time-consuming and expensive. But with growing interest—and $90 million over 5 years from the NIH Common Fund initiative (see the sidebar)—there is cautious optimism that over the next decade single-cell research may begin to pay dividends for patients with cancer and other diseases.
Moving beyond the Average
Most research on the molecular biology of tumors requires the use of mixtures of tens or hundreds of thousands of cells. Those samples “have immune cells, endothelial cells, and other infiltrating cells that make up the milieu of what a tumor actually is,” explained Dr. Dan Gallahan of NCI’s Division of Cancer Biology. “That really makes it difficult to get a grasp on what defines or, more importantly, how to treat a tumor.”
Results from studies that involve a bulk population of cells, Dr. Gallahan continued, essentially represent an average measurement.
Studying single cells is a way to “defy the average,” Dr. Marc Unger, chief scientific officer of Fluidigm Corporation, said earlier this year at a stem cell conference in Japan. (Fludigm, which develops tools for single-cell analysis, and the Broad Institute recently announced plans to establish a single-cell genomics research center.)
Single-cell analysis may be able to provide important clinical insights, said Dr. Nicholas Navin of the University of Texas MD Anderson Cancer Center, who has used next-generation sequencing to study variations in the number of genes (copy number variation) in single cancer cells.
Thanks to technological advances, a growing number of investigators are beginning to delve deeper into the biology of the single cell.
Single-cell analysis might, for example, help identify “pre-existing [cell populations] that are resistant to chemotherapy or rare subpopulations that are capable of invasion and metastasis,” he said. “We may also be able to quantify the extent of heterogeneity in a patient's tumor using single-cell data and use this index to predict how a patient will respond to treatment,” Dr. Navin continued.
Results from several recent studies have highlighted the challenges posed by tumor heterogeneity.
For example, researchers at BGI (formerly Beijing Genomics Institute) sequenced the protein-coding regions of DNA (the exome) of 20 cancer cells and 5 normal cells from a man with metastatic kidney cancer. The researchers found a tremendous amount of genetic diversity across the cancer cells, with very few sharing any common genetic mutations.
Much of the work in the analysis of single cells is still quite preliminary, and any potential clinical impact is still some years away, researchers agree.
“The problem with the single-cell data is that we don't really know yet what they mean,” Dr. Sangeeta Bhatia of the Massachusetts Institute of Technology commented recently in Nature Biotechnology.
And studies involving bulk populations of cells will not be going away any time soon, noted Dr. Betsy Wilder, director of the NIH Office of Strategic Coordination, which oversees the NIH Common Fund.
“Single-cell analysis isn’t warranted for every question that’s out there,” Dr .Wilder stressed. “Studies using populations of cells will continue to be done, because it makes a lot of sense to do them.”
Technology, a Driving Force
Beyond just an interest in learning more about single cells—what Dr. Gallahan called “the operational units in biology”—technology has been the driving force behind the growth of this field.
Dr. Stephen Quake, also of Stanford, has pioneered the use of microfluidics, which typically uses small chips with microscopic channels and valves—often called lab-on-a-chip devices—that allow researchers to single out and study individual cells. Dr. Quake, who co-founded Fluidigm, and others are increasingly using these devices for gene-expression profiling and for sequencing RNA and DNA of individual cells.
Dr. Nolan’s research involves a hybrid approach that combines two technologies: a souped-up method of flow cytometry, which has been used for several decades to sort cells and to perform limited analyses of single cells, and mass spectrometry, which is often used to identify and quantify proteins in biological samples.
Dr. Nolan’s lab is using this “mass cytometry” approach—developed by Dr. Scott Tanner of the University of Toronto—to characterize the response of individual cells to different stimuli, such as cytokines, growth factors, and a variety of drugs. Much of the group’s work has focused on analyzing normal blood-forming cells.
They published an influential study last year in Science that revealed some of the subtle biochemical changes that occur during cell differentiation. The study also described how dasatinib (Sprycel), a drug used to treat chronic myelogenous leukemia, affects certain intracellular activities. The research, Dr. Nolan said, is a prelude to studying individual cells from patients with blood cancers. The approach, he believes, may prove particularly useful for identifying new drugs and for testing them in the lab.
A tumor is not a uniform mass of identical cells with identical behaviors. Cells can act quite differently in one part of a tumor than in another.
The Microscale Life Sciences Center (MLSC), an NIH Center of Excellence in Genomic Science that is housed at Arizona State University, develops and applies the latest technology to single-cell research.
The center—a collaboration of investigators from Arizona State, the University of Washington, Brandeis University, and the Fred Hutchinson Cancer Research Center—includes researchers from numerous disciplines, including microfluidics, computer science, physics, engineering, and biochemistry, explained principal investigator Dr. Deirdre Meldrum.
“All of these disciplines are needed to develop the new technologies we’re working on,” said Dr. Meldrum, an electrical engineer by training.
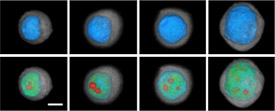
In its initial work, the MLSC has measured metabolic processes in single living cells, including cellular respiration—the process by which cells acquire energy—as it relates to an individual cell’s ability to resist or succumb to cell death. The workhorse of this effort is a platform called the Cellarium, developed by Dr. Meldrum’s team. Individual cells are isolated in controlled chambers, Dr. Meldrum explained, “where we perturb them and measure how they change over time.”
Investigators at the MLSC and elsewhere have also developed technologies to image single cells. MLSC scientists are using a device developed by VisionGate, called the Cell-CT, “that enables accurate measurement of cellular features in true 3D,” Dr. Meldrum said.
MLSC researchers have studied abnormal esophageal cells from people with Barrett esophagus, a condition that increases the risk of esophageal adenocarcinoma. In particular, they’ve looked at how these cells respond to very low oxygen levels, or hypoxia.
Acid reflux, which can cause Barrett esophagus, can damage the esophagus “and lead to transient hypoxia in the epithelial lining of the esophagus,” explained Dr. Thomas Paulson, an investigator at Fred Hutchinson. In effect, he continued, the Cellarium system provides a snapshot of how this hypoxic environment selects for variants of cells that are able to survive and grow in it, providing insights into the factors that influence the evolution of cells from normal to cancerous.
Although Dr. Paulson’s work at MLSC is focused on Barrett esophagus, he believes the approach represents an excellent model system for studying cancer risk in general.
“I think our understanding of what constitutes risk is probably going to change as we understand the types of changes that occur at the single-cell level” that can transform a healthy cell into a cancerous cell, he said.
Deeper Dives Ahead
There’s a general acknowledgement in the field that single-cell analysis still has important limitations. Technological improvements are needed that can allow for the same type of molecular and structural “deep dives” that can be achieved by studying batches of cells. And powerful computer programs will be needed to help interpret the data from single-cell studies.
In addition, the research will eventually have to move beyond the confines of the mostly artificial environments in which single cells are now being tested, Dr. Gallahan noted. “As the technology gets better, we should be able to do more of this work in an in vivo setting.”
Although much more work is needed, the potential for what can be learned from studying single cells is quite large, Dr. Nolan believes.
“The fact that we’ve been able to make good decisions and learn as much as we have, even at the level of resolution [of cell populations], means that there’s something of even greater value to mine when you get to the level of the single cell,” he said.
—Carmen Phillips
Transforming the Field of Single-Cell Research
This month, the National Institutes of Health will announce grant recipients for the NIH Common Fund’s single-cell analysis program.
The program, which includes three funding opportunities, “is largely a technology building program,” explained Dr. Wilder. The NIH Common Fund launched this program now because “there’s a sense that the technologies exist that can enable us to do the sort of analysis required to look at single cells in their native environment,” such as in a piece of excised tissue.
Although the focus is on technology, an important goal of the initiative is to support research that will “identify a few general principles of how single cells behave in a complex environment,” added Dr. Ravi Basavappa, the program director for the single-cell analysis program.
From the planning discussions, it was clear that the program should not limit the types of technology under consideration, Dr. Wilder commented. “Our analysis indicated that there are a lot of possibilities, so we left it up to the imaginations of the investigators to determine what technologies would be most transformative for the field as a whole.”