New Directions in Biology and Medicine - AAAS Plenary Lecture
Statement of
Harold Varmus, M.D.
Director, National Institutes of Health
Plenary Lecture, American Association for the Advancement of Science
Philadelphia
February 13, 1998
- Today the President of the United States came to Philadelphia to celebrate science and this organization's 150th anniversary;
- two weeks ago in his State of the Union Address, he spoke forcefully and at length about the importance of science and education in our society and proposed a new Research Fund for America and the 21st Century;
- then, six days later, he asked Congress for the largest increase in research funding in history--not just a one year increase, but an increase sustained over five years;
- and, during the past several months, many members of Congress, on both sides of the aisle, have also been calling for major expansions of our Federal research programs, also over the next five or ten years.
For those of us who came to Washington four years ago, amidst doubts that the Federal budget could ever be balanced and amidst warnings that funding for science would soon decline, these events seem miraculous. But they are true.
In his address earlier today, the President outlined his aspirations for science and his views on our new circumstances. My goal is to offer a medical scientist's perspective on our change of fortunes, to illustrate graphically some of the science that excites us, and to describe some of the benefits our science can deliver to society.
My thesis is simple: the timing of this change could not be better. New technologies and new knowledge have revolutionized our abilities to understand normal biological functions and disease. A broad array of scientific disciplines made this revolution, and that consortium is now required more than ever if we are to follow the new paths. A national commitment to train and support the best minds in these varied disciplines can improve the health of people here and throughout the world. Expanded funding, exciting science, and public benefit--this is a package that should make everyone cheer.
How do we make the most of this opportunity? I will try to answer this question for my own galaxy in the universe of science by discussing four broad ideas. These are based on my view of the recent history and current state of biomedical science. They will be supported by some pictures of provocative science and embroidered by some opinions about science policy.
Theme #1: Discoveries in biology and medicine depend on progress in many fields of science.
Most of the revolutionary changes that have occurred in biology and medicine are rooted in new methods. Those, in turn, are usually rooted in fundamental discoveries in many different fields. Some of these are so obvious that we lose sight of them--like the role of nuclear physics in producing the radioisotopes essential for most of modern medical science. Physics also supplied the ingredients fundamental to many common clinical practices--X rays, CAT scans, fiber optic viewing, laser surgery, ECHO cardiography and fetal sonograms. Materials science is helping with new joints, heart valves, and other tissue mimetics. Likewise, an understanding of nuclear magnetic resonance and positron emissions was required for the imaging experiments that allow us to follow the location and timing of brain activities that accompany thought, motion, sensation, speech, or drug use. Similarly, X-ray crystallography, chemistry, and computer modeling are now being used to improve the design of drugs, based on three-dimensional protein structures.
The Human Genome Project, which is now churning out the maps and nucleotide sequences of the chromosomes of many organisms, from microbes to man, would not exist without recombinant DNA methods; molecular cloning, in turn, would not exist without earlier studies of enzymes for synthesizing, cutting, and repasting DNA. Moreover, today's efforts to complete the 3 billion base sequence of human DNA by 2005--and the sequence of many other genomes as well--could not operate in high gear without robots for processing many samples and, more importantly, without computers to manage, store, compare, and retrieve the data.
Recent efforts to sequence DNA on a commercial scale--for example, to screen many individuals for mutations that predispose to certain cancers--use nanotechnology and photochemistry to synthesize arrays of nearly 100,000 different short pieces of DNA on a small chip. DNA tagged with a fluorescent probe can then be added to the chip to determine the sequence of a certain gene, such as BRCA-1, a gene that influences the likelihood of breast and ovarian cancer. In an example recently published by investigators from the Affymatrix Corporation and the National Human Genome Research Institute (NHGRI), a patient's DNA tagged with a red fluor was compared with a normal DNA sample tagged with green to seek mutations in part of the BRCA-1 gene. In the magnified version  , the red square indicates the position of a mutation in the patient's genome that predicts a high likelihood of cancer.
, the red square indicates the position of a mutation in the patient's genome that predicts a high likelihood of cancer.
An even more colorful new method developed by Thomas Reid and his colleagues at the NHGRI, spectral karyotyping, uses chromosome-specific fluorescent probes, sophisticated optics, and computer-based analysis of the complex emissions to paint each of the 24 types of human chromosome with a different color. This allows (for example, as shown at the bottom of the aaas-figure) swift identification of rearranged chromosomes in different human cancers.
These are but few of many examples of the dependence of biomedical sciences on a wide range of disciplines--physics,  chemistry, engineering and many allied fields. These examples underscore the need to sustain the strength of our sciences across the full spectrum, not just protect the popular health sciences, which have fared relatively well in the past three rounds of Congressional appropriations. For this reason, those of us in the health sciences applaud the breadth of the President's proposal--and the bipartisan proposals from many in Congress--to expand funding for Federal science agencies supporting diverse disciplines.
chemistry, engineering and many allied fields. These examples underscore the need to sustain the strength of our sciences across the full spectrum, not just protect the popular health sciences, which have fared relatively well in the past three rounds of Congressional appropriations. For this reason, those of us in the health sciences applaud the breadth of the President's proposal--and the bipartisan proposals from many in Congress--to expand funding for Federal science agencies supporting diverse disciplines.
Theme #2: Methods that dramatically expand biological data also demand new modes of analysis and new ways to ask scientific questions.
Two months ago, the President awarded the National Medal of Technology to three people whose work anticipated and ultimately shaped the way biology and medicine are studied in this new era. As long ago as the1950's and 1960's, Robert Ledley was describing how medical information could be stored and analyzed with computers. In 1964 for instance, he wrote in Science magazine that the many photographs being taken of biological phenomena could be analyzed by digital computers to avoid "the tedium. . .precision, and extensive time" required by manual methods. In the 1970's, Vint Cerf and Robert Kahn were inventing the platforms for the Internet, the system that now seems indispensable to scientists of all kinds who use it to talk with each other, exchange new results, explore data bases, and seek new technologies.
The visionary activities of these three computer scientists illustrate vividly how one science can profoundly alter the way another science is done. Every day, post-docs in my lab log on to the NIH GenBank or other sites, such as the Human Transcript Map, to see if someone working on any organism--yeast, flies, or man--may have discovered something about a gene we are studying in mice. At such times, computer science and genetics seem consanguineous. Two virtues of computers--interconnectedness and interoperability--seem to mirror two great truths revealed by modern genetics: the universality of the genetic code (that allows us to read genetic information in all species) and the evolutionary conservation of genes (that allows us to predict what they do in one organism by studying another).
Computers permit all biologists to contemplate vastly greater amounts of information than they were able to think about before. At the same time, new experimental methods allow single investigators to gather much more data than was possible a decade or two ago.
I have seen this most clearly in my own field of cancer biology. In the 1970's and 1980's, it was usual for a laboratory group like mine to focus on one or two genes suspected to have a role in animal or human cancer. We would isolate the gene, study its protein product, and try to discern the function of the protein and its location in cells--like the protein kinase encoded by the src oncogene. This mode of science helped to establish important principles--for example, that cancer arises from mutations in normal cellular genes--but could not tell the full story about cancer or about control of normal growth.
Now new methods make it possible to generate much more complex pictures of genetic damage and gene activity in cancer cells. For this reason, the National Cancer Institute and the National Library of Medicine have recently launched the Cancer Genome Anatomy Project (CGAP), available to all of you through the Internet. Samples of tumors--and of normal tissue from which the tumors arose--are obtained by laser dissection, then studied to determine which genes are expressed and which are not, and to compare levels of expression in normal and malignant tissue.
One especially potent method for doing this is the expression array  . Fragments of many isolated genes are arrayed on a small piece of glass or nylon, then challenged with fluorescent-labeled DNAs that reflect the levels of expression of individual genes in the test cells. If one sample is labeled green and one sample red, it is possible to compare levels of expression in the two cell populations by recording the intensity of emissions at two wave lengths. For the viewer, the color holds the message: yellow means no difference, red or green means the gene is expressed at different efficiencies in the two samples.
. Fragments of many isolated genes are arrayed on a small piece of glass or nylon, then challenged with fluorescent-labeled DNAs that reflect the levels of expression of individual genes in the test cells. If one sample is labeled green and one sample red, it is possible to compare levels of expression in the two cell populations by recording the intensity of emissions at two wave lengths. For the viewer, the color holds the message: yellow means no difference, red or green means the gene is expressed at different efficiencies in the two samples.
Although we do not yet have all 80,000 human genes in hand to measure with this method, we can follow the activity of hundreds and even thousands. A recent example from the labs of Lou Staudt at the NCI and Pat Brown at Stanford  compares an indolent and malignant form of a lymphoma and demonstrates differences in expression of many genes, including some that could affect the choice of chemotherapy.
compares an indolent and malignant form of a lymphoma and demonstrates differences in expression of many genes, including some that could affect the choice of chemotherapy.
The power of this revolutionary method is most graphically represented in experiments with the simple eukaryotic organism, baker's yeast. The complete sequence of yeast DNA has been determined by an international consortium, and all of its 6100 genes can be arrayed on a single small glass slide. The activity of all of the organism's genes can then be examined as the yeast are exposed to a variety of environmental signals. For example, as portrayed from work in Pat Brown's lab at Stanford in  , when yeast are grown in glucose until it is depleted, they must suddenly switch to ethanol for nourishment. At that point, there is wild fluctuation in gene activity, as evident from the many red and green spots on this quadrant of the complete array. How can we make sense of these many changes? First, some clever software can sort the genes into cohorts that show the same pattern of changes. Second, genes with functions known to be related can be found within these cohorts
, when yeast are grown in glucose until it is depleted, they must suddenly switch to ethanol for nourishment. At that point, there is wild fluctuation in gene activity, as evident from the many red and green spots on this quadrant of the complete array. How can we make sense of these many changes? First, some clever software can sort the genes into cohorts that show the same pattern of changes. Second, genes with functions known to be related can be found within these cohorts 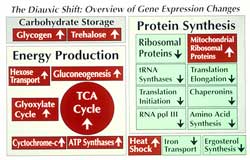 , offering insight--and provoking speculation--about the physiological meaning of the changes. Finally, we will have to learn to test those speculations.
, offering insight--and provoking speculation--about the physiological meaning of the changes. Finally, we will have to learn to test those speculations.
Learning to do this--for yeast or mammals--will not be easy. For two decades we have been discovering proteins, like the src kinase, involved in growth control and cancer in mammalian cells. But we now know that there are many interacting pathways that transmit growth signals from the cell surface to the nucleus, as summarized by Tony Hunter of the Salk Institute in  . These pathways have come to seem truly bewildering, even to those who study them. Here is a place where experimental biologists need help--perhaps from mathematicians or chaos theorists or engineers.
. These pathways have come to seem truly bewildering, even to those who study them. Here is a place where experimental biologists need help--perhaps from mathematicians or chaos theorists or engineers.
To promote such interdisciplinary work, the NSF and the NIH have recently agreed to sponsor a program that will allow scientists from many spheres to study biological problems in the NIH intramural research program. We are also fostering new efforts in materials science, bioengineering, instrumentation development, and informatics.
In short, biology is not only for biologists.
Theme #3: Genetics, biochemistry, and structural biology are accelerating a transition from medical serendipity to rational, mechanism-based control of disease.
Skeptics have argued that my colleagues and I are overly excited about new genes and powerful molecular methods; that this new knowledge has not been effectively harnessed to prevent or treat disease; and that the staples of medicine are still the consequences of good luck or random screening (like penicillin or digitalis or the current cancer drugs). I think the skeptics are merely impatient.
Consider these results, presented at last week's Retrovirus meetings in Chicago  . The sharply downward slope shows that recently developed antiviral drugs actually reduce AIDS mortality rates. We know these drugs do not cure patients and are far from perfect--they are expensive, sometimes poorly tolerated, and ineffective against certain variants of HIV. But they have enormous symbolic as well as practical significance: They are harbingers of an age in which medicine will be based on a detailed understanding of the genetic and biochemical mechanisms of disease.
. The sharply downward slope shows that recently developed antiviral drugs actually reduce AIDS mortality rates. We know these drugs do not cure patients and are far from perfect--they are expensive, sometimes poorly tolerated, and ineffective against certain variants of HIV. But they have enormous symbolic as well as practical significance: They are harbingers of an age in which medicine will be based on a detailed understanding of the genetic and biochemical mechanisms of disease.
In the case of AIDS, we know that levels of virus in the blood and thus the rate of viral production predict the progress of disease. Dissection of the genomes of many retroviruses, including the genome of HIV, has taught us that these viruses encode and absolutely require at least three enzymes for successful multiplication: a reverse transcriptase to make viral DNA; an integrase to join the DNA to chromosomes, and a protease to carve functional viral proteins out of larger precursors. Effective inhibitors have been made against two of these enzymes (reverse transcriptase and, as shown on the slide, protease). These drugs reduce virus levels rapidly and profoundly in infected people, especially when used in combination. This is the formula that has so dramatically reduced AIDS mortality rates.
The ingredients of this partial victory against AIDS will, I believe, be fundamental to success in other venues. The rules are simple: Know the disease process at the cellular level. Identify the responsible genes and their protein products. Focus on these molecules to prevent or revert the disease with new vaccines, drugs, or other approaches.
This recipe has been effective in the past for ailments other than AIDS: treatment of endocrine disorders by hormone replacement; prevention of hepatitis B virus infection by vaccination with a recombinant protein; reduced death rates from coronary artery disease by drugs that interfere with cholesterol metabolism. But the greatest triumphs lie ahead--against infectious diseases, metabolic diseases, neurological diseases, and cancer.
The near-term prospects for advances against cancer seem especially strong. They are reflected in the emphasis the President and Vice-President have placed on cancer in their recent budget proposal, and they were summarized by the President in his speech here today. Even with blunt tools, we have made significant progress; we can now cure most pediatric and a few adult cancers that were once uniformly lethal; we know that many cancers can be prevented by avoiding the use of tobacco products; and we have just observed a decline in overall age-adjusted mortality rates for the first time in history.
But what lies ahead is much greater and depends on many of the advances I have described here today. For example:
- Expression arrays can be used to classify cancers more rigorously, allowing more informed choice of therapy.
- Knowledge of genes deranged in cancer offers many new targets for treatment. We can now think creatively about ways to interfere with signals that tell cancer cells to grow and divide, to block the blood vessels that feed a tumor, and to protect normal cells from drug toxicity.
- Sequencing chips can identify mutations that predispose to many cancers, including several kinds, like skin and intestinal cancers, that we need to be warned about--because they can now be detected at very early stages.
- And better methods for visualizing small tumors in the brain, breast, and other organs offer the prospect of diagnosis at a stage when surgical cures are more likely.
As we contemplate these improvements, however, we must also remember a couple of sobering facts. First, clinical advances require clinical research, an embattled enterprise that needs help to recruit, train, and support talented people. Second, the pace of progress in the clinical arena is different from the heady pace in today's laboratories; it is measured in years or decades, not months and days.
Theme #4: Advances in biology and medicine in the developed world can and must be used to combat disease in the developing world.
The trends in biology and medicine that I have described are occurring mainly in the most advanced nations and being applied to the health problems encountered in those countries. Of course, many of these problems are shared with poorer, less developed countries. This is well-known for AIDS because of the magnitude of the AIDS epidemic in Africa and Asia. It is also becoming true of most of the diseases associated with aging, because demographers are showing us that the percent increases of people over 65 will soon be greater in some of the least developed countries, like Indonesia, than in the developed nations. Nevertheless, there is a real danger that biomedical research will not reach its potential in such places because the solutions we find for our citizens may not be affordable where average annual incomes are less than one thousand dollars and where average health care expenditures per person are less than one percent of ours. Furthermore, some of the greatest health threats--especially certain infectious diseases--are largely confined to the developing world. But there are often inviting targets for biomedical science.
Consider malaria, a disease that seemed to be vanishing in the 1950's but is now resurgent in many parts of the world due to failed insect control programs and resistance of the causative agent (the protozoon parasite called Plasmodium) to conventional drugs. Infection with this organism affects over 300 million people each year  , and about 2 million people die, mainly young children and pregnant women in sub-Saharan Africa. Nevertheless, the world's scientific community devotes only about $100 million to malaria research each year, and the drug industry has limited interest. But the prospects seem enormous for designing new interventions with modern tools of cell and molecular biology. The organization of a Plasmodium genome project and the recent isolation of the gene conferring chloroquine resistance in strains now spread around the world should enhance the possibilities of more effective drugs and new vaccine candidates.
, and about 2 million people die, mainly young children and pregnant women in sub-Saharan Africa. Nevertheless, the world's scientific community devotes only about $100 million to malaria research each year, and the drug industry has limited interest. But the prospects seem enormous for designing new interventions with modern tools of cell and molecular biology. The organization of a Plasmodium genome project and the recent isolation of the gene conferring chloroquine resistance in strains now spread around the world should enhance the possibilities of more effective drugs and new vaccine candidates.
Some of us believe that research efforts on diseases like malaria need to extend well beyond what can be accomplished in laboratories in the advanced countries. For this reason, agencies in several countries, in conjunction with the World Bank and World Health Organization, have formed a Multi-lateral Initiative on Malaria to promote collaborative research, conducted--at least in part--in Africa. The immediate goal is to improve African research capabilities; this is already happening through the support of multi-center projects and through establishment of Internet accessibility at several research centers. But we envision many long-range benefits as well: the cultural pride associated with such centers, the opportunity to inspire African students to enter research careers, and the ultimate improvements in health that can help stabilize developing countries politically and economically.
My own enthusiasm for this venture is based, in part, on a recent visit to the U.S.-supported Malaria Research Center in Bamako, Mali  . There, NIH-trained and -funded African scientists study plasmodial drug resistance and distribution of Anopheles mosquitos with sophisticated genetic methods; map epidemiological patterns with Global Positioning System devices; and keep in touch with the rest of the world through computer connections established by the NIH's National Library of Medicine. The Malaria Center draws American scientists to Bamako, sends investigators to villages to control infection, and wins the attention and approbation of government and business leaders.
. There, NIH-trained and -funded African scientists study plasmodial drug resistance and distribution of Anopheles mosquitos with sophisticated genetic methods; map epidemiological patterns with Global Positioning System devices; and keep in touch with the rest of the world through computer connections established by the NIH's National Library of Medicine. The Malaria Center draws American scientists to Bamako, sends investigators to villages to control infection, and wins the attention and approbation of government and business leaders.
If this can happen in one of the poorest countries in the world, it can happen anywhere. One requirement is the international resolve to use our science for goals that affect us only secondarily--through the enhanced well-being and stability of other nations. We also need our own trainees to look beyond the research activities of their mentors and seek some of the daring opportunities that the new biology offers to combat malaria, tuberculosis, and other relatively neglected problems.
In the few minutes we have had together this evening--and with a few examples chosen from the many possible--I have tried to convey my sense that biology is being transformed. This transformation is driven by new methods--methods that allow us to survey the genetic landscape of all living things; to envision the shape and function of molecules, cells, and organs; and to change the behavior of biological systems through genetics and chemistry. Implicit in what I have said are notions that traditionally drive public investment in science: that knowledge, itself wonderful and ennobling, is also useful for society. I have tried to portray the usefulness of biology in the field I know best--the battle against disease, in countries both rich and poor. But for other biologists, the utility of our science can mean many things that I have not discussed--improved food production, environmental protection, or industrial strength.
For each of these goals, social usefulness is inevitably tied to ethical and legal concerns. In modern medical science, these concerns are now legion and widely publicized: How do we protect against abuses of genetic information? How do we properly design and oversee clinical trials? How do we balance reproductive rights against our responsibilities to our genetic lineage? Of course, I cannot answer such questions this evening, but I do want to make a simple point about who should be responsible for answering them: Just as biology is not just for biologists, so are ethics and law not just for ethicists and lawyers.
An example: This week the Senate briefly debated an anti-cloning bill that would have also outlawed the use of so-called somatic cell nuclear transfer to develop cell-based therapies for human diseases. Efforts to bring the bill to a swift vote without the usual deliberative process were defeated by a bipartisan action in which scientists and patient advocacy groups played an indispensible role--the role of educating legislators about the important science the bill would restrict. Senator Connie Mack made my point very clearly in yesterday New York Times' account of these events: "Let's hear from the scientific community," he said, " so they can tell us whether this is the right thing to do or the wrong thing to do."
Now neither Senator Mack nor I want to turn the final decisions over to the scientific community. What I am suggesting is the need for an expanded role for scientists in the public debate. Traditionally, we have been appropriately concerned about the scientific literacy of those engaged in such debates. But we need to go beyond this--to encourage scientists to become involved themselves in the social, legal, and ethical issues their work provokes.
How do we do this? The NIH, mainly through the NHGRI, has for several years been supporting research on the societal dilemmas posed by genetics. Recently, we have brought all of the Institutes into discussions of a wide range of ethical and legal questions raised by biomedical research. But I believe that we need to start these discussions much earlier--during the process of becoming a scientist.
To this end, I would like to offer a modest challenge to the AAAS. Every week, Science magazine reports very effectively about social and ethical consequences of contemporary science. Why not cull several interesting stories from weekly issues of Science to assemble a slim monthly magazine for distribution, free of charge, to high school science classes and college students? I believe that a magazine of this type would be popular, would alert science-oriented students to the potential impact of their future work, and might even attract students oriented elsewhere to rethink career goals.
In his State of the Union message two weeks ago, the President's endorsement of science was followed by his conviction that "we must continue to see that science serves humanity, not the other way around." As we applaud his commitment and celebrate our achievements, let us also embrace our responsibilities.
Thank you and good night.


