dbVar Help
- Introduction
- dbVar Study Browser
- dbVar Study page
- dbVar Variant page
- dbVar Placements
- FTP site
- dbVar Entrez search
Last updated: October 25, 2011.
Introduction
dbVar is a database of genomic structural variation that allows you to search, view, and download variant data from studies submitted for any organism. In general, variants are ≥ 50 nucleotides, but are occasionally smaller. dbVar provides access to the raw data (when available) and links to other NCBI and external resources. For more information on structural variation in general see the Overview of Structural Variation page and for frequently asked questions about dbVar see the dbVar FAQ page.
dbVar is a free resource that is developed and maintained by the National Center for Biotechnology Information (NCBI), at the U.S. National Library of Medicine (NLM), located at the National Institutes of Health (NIH) in Bethesda, Maryland.
dbVar Study Browser
Data in dbVar is organized by Study. "Study" typically refers to a publication, however some Studies are community resources which are updated with new data on a regular basis (e.g., International Standards for Cytogenomic Arrays - ISCA, nstd37). Each Study is given an accession that begins with '*std': nstd if the study was submitted to NCBI and estd if the study was submitted to EBI. The dbVar Study Browser (Figure 1) provides a summary of the studies in dbVar. The browser can be sorted by Study accession, Organism, Study Type, Method, Number of Variant Regions, Number of Variant Calls, or Publication. Links to the Study Page and PubMed summary for each study are provided. Filters to the right of the browser allow you to narrow content by Organism, Study Type, Method, or Number of Variant Regions.
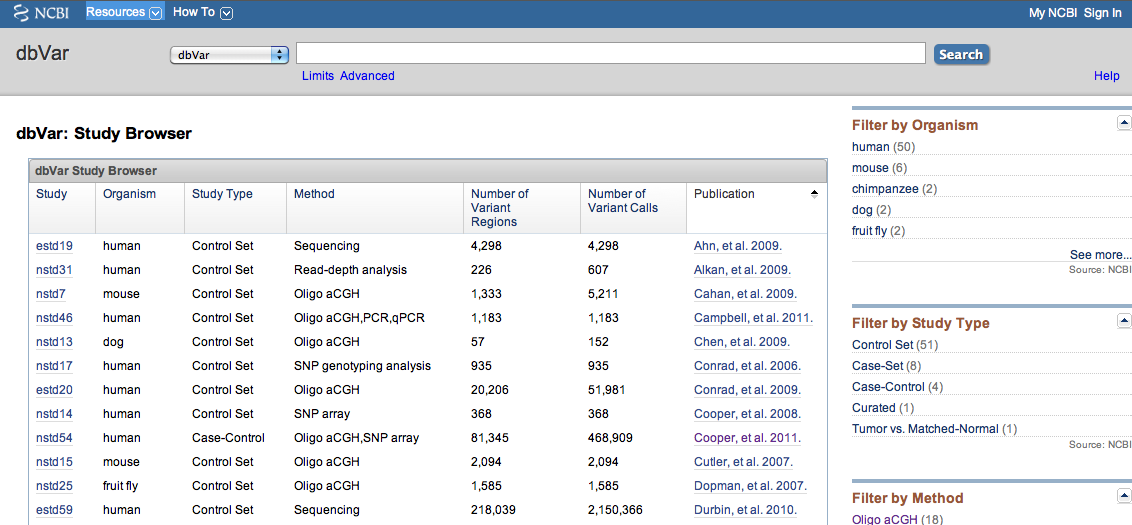
Figure 1: dbVar Study Browser
dbVar Study page
Each Study in dbVar has a Study Page that displays basic information about the study. At the top is a general information section describing basic information about the study, including links to BioProjects, PubMed, dbGaP, and the submitter's lab page (if one was provided). This is followed by a Detailed Information section where you can download variant data for the current study (Variant Regions, Variant Calls, Both, or everything via FTP) or browse details about Variant Summary, Samplesets, Experimental Details, or Validations in a tabbed format.
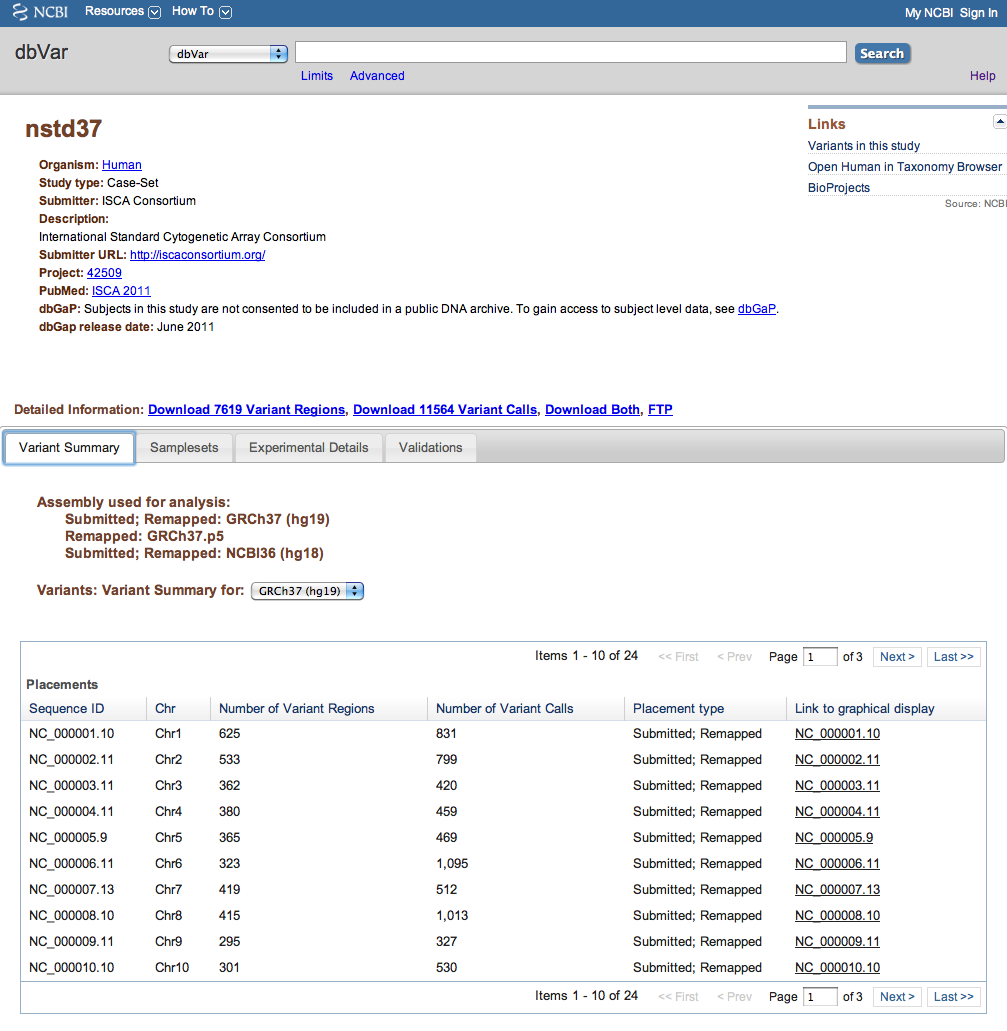
Figure 2a: General Information and Variant Summary Tab (Study Page) - The Variant Summary tab displays the number of Variant Calls and Variant Regions for the current study on each chromosome, Placement type (i.e., whether data is available on the Submitted assembly and/or Remapped assemblies), and links to a graphical display of the variants on NCBI's Sequence Viewer. A pull-down menu above the table allows you to display the current study's data on the submitted assembly, or on any assemblies to which the data has been remapped.
Figure 2b: Samplesets Tab (Study Page) - The Samplesets tab describes any logical division of samples in the study. Details include Name, Description, Size, and relevant Phenotypes, if any. Note that additional details about individual subjects in a sampleset can be found by downloading the Samples data. If subjects have not been consented to have their data displayed publicly (as in the example above), clinical information and links between genetic variants are stored behind controlled access at NCBI's dbGaP. Samples and variants are then anonymized before being forwarded to dbVar.
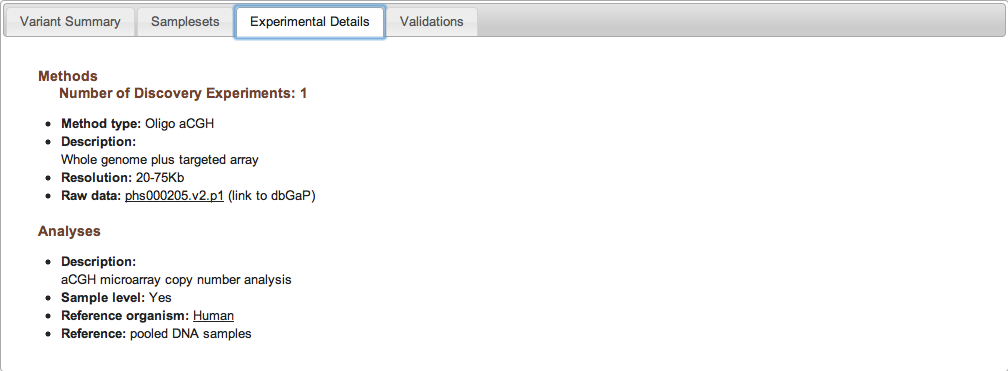
Figure 2c: Experimental Details Tab (Study Page) - The Experimental Details tab provides information on the methods and analyses that were used in the study. Each unique combination of method and analysis is named an "Experiment." A complete list of allowable Method Types and Analysis Types can be found in the Excel submission template, or by clicking here. Experiments can be one of three types: Discovery, Validation, or Genotyping. The Experimental Details tab is also where you can access links to the study's raw data (e.g., sequence traces or array data stored in an external database such as Trace or Array Express).
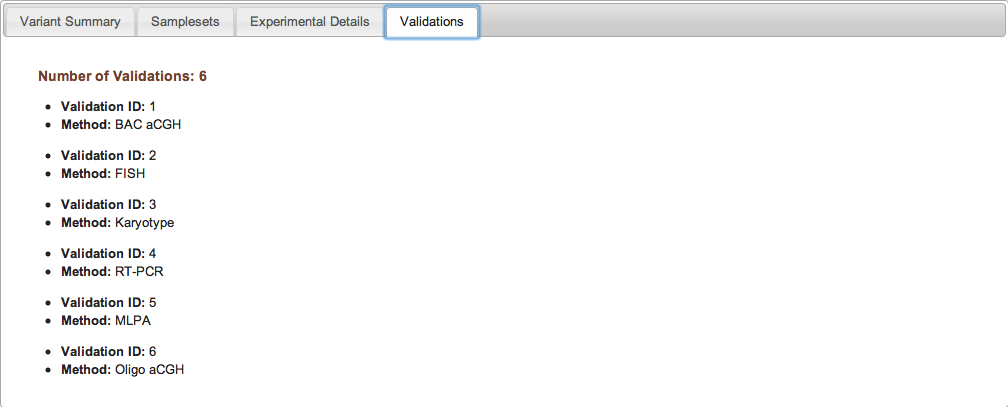
Figure 2d: Validations Tab (Study Page) - If validation experiments were performed to confirm variant calls generated by discovery experiments, the validation experiments are assigned unique IDs and are listed along with the relevant method in the Validations tab. Individual validation results can be accessed by downloading the variant data itself from the Study Page. If no validation experiments were performed for a given study, the Validations tab will indicate that no validation data were submitted to dbVar.
dbVar Variant Page
Each dbVar Variant page displays detailed information about an accessioned Variant Region. At the top of the page is a section with general information about the Variant Region (Organism, Study, number of supporting Variant Calls, Region size, etc. - see Figure 3a), and a chromosome ideogram displaying the variant's location on the genome. This is followed by a tabbed section with complete variant details: Genome View, Variant Region Details and Evidence, Validation Information, Clinical Assertions, and Genotype Information (see figure legends below for information on each of these tabs).
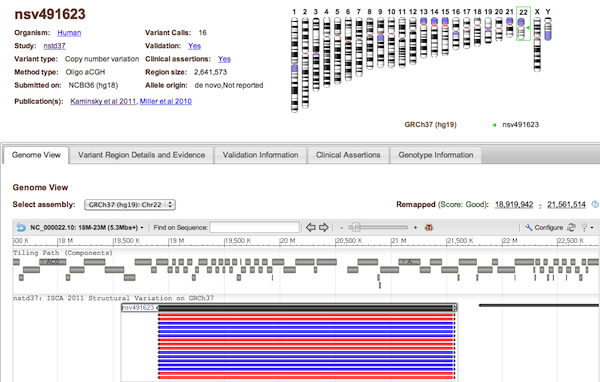
Figure 3a: General Information (top) and Genome View Tab (Variant Page) - The Genome View Tab shows the current variant in NCBI’s Sequence Viewer in the context of known genes and other variant data from the current study. Use the pull-down menu to select the assembly you want to view. Each black bar represents a Variant Region (the current region is centered and highlighted), while the colored bars represent Variant Calls that support each region. The display default is to collapse all supporting variant calls, but this can be adjusted (for example, to show all individual calls) by clicking Configure… Variations… Show All. Clicking on any Variant Region or Variant Call will reveal a pop-up tooltip with information about the variant and a link to its dbVar page.
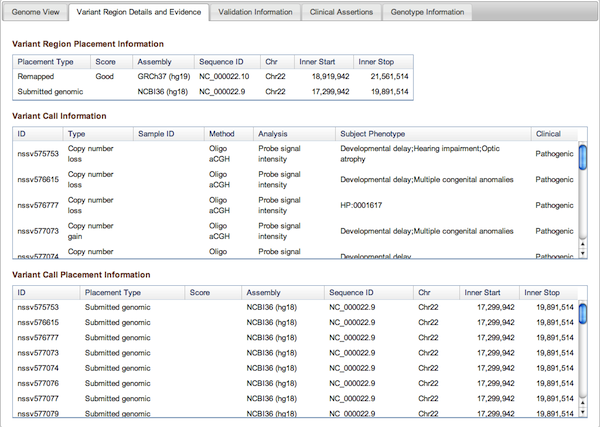
Figure 3b: Variant Region Details and Evidence Tab (Variant Page) - This tab contains placement coordinates for the variant region on the assembly on which the region was originally submitted as well as assemblies to which it was subsequently mapped. In the case of remapped placements, the Score column indicates the quality of the remap – Perfect, Good, or Pass (for details see dbVar Placements section below). Below the Variant Region information are details on the supporting Variant Calls that were merged to define the Region, including their placements (on Submitted and/or Remapped assemblies). Complete details can always be downloaded using the links above the tabbed section of the Variant Page.
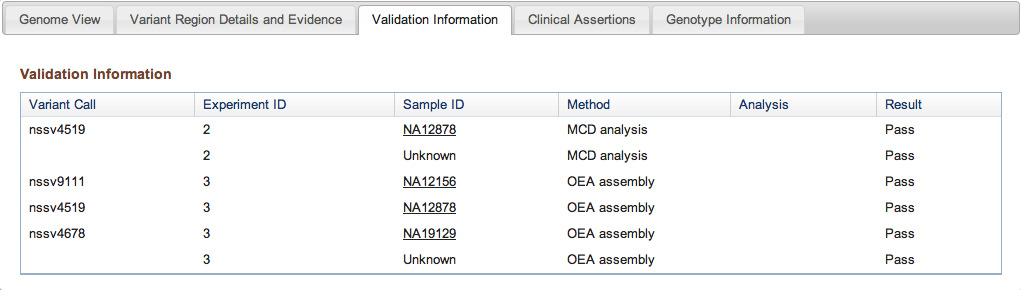
Figure 3c: Validation Information Tab (Variant Page) - If any Variant Call results were validated with additional methods, this tab provides details on the methods and analyses used in the validation, and the results (Pass or Fail for each call tested).
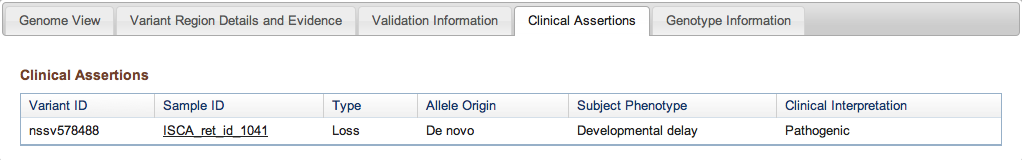
Figure 3d: Clinical Assertions Tab (Variant Page) - If in the course of a study an association was established between a Variant Call and a phenotype observed in a Subject, details are provided here. The Variant Call ID is followed by the sample in which it was observed (with a link if the sample is publicly available), the type of event (insertion, deletion, copy number gain, etc.), the parental origin of the variant if it was determined, the phenotype associated with the variant, and the authors’ assessment of its likely pathogenicity. For a reminder of how studies involving sensitive clinical and genetic information are processed, please refer to the Figure 2b legend above.
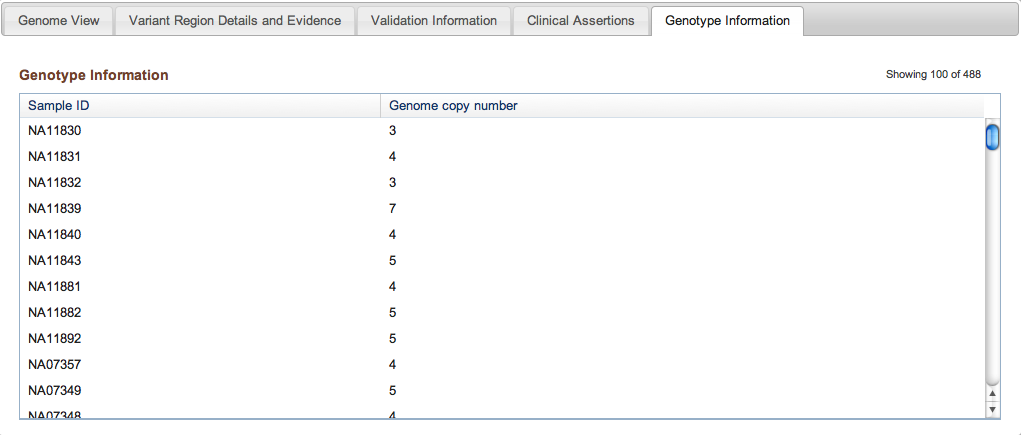
Figure 3e: Genotype Information Tab (Variant Page) - Lastly, the Genotype Information tab contains any genotype data that were submitted as part of the study. Genotype data are generated by a separate Experiment (see above), and can reflect the investigation of Discovery variants from the current study, or existing variants from another published study (in which case the dbVar accession for the existing variant must be included with the submission). NCBI is currently developing a Genotype Server that will eventually collect and archive genotype data on all structural variation and SNP data in a centralized location.
dbVar Placements
Most variants are submitted to dbVar as asserted locations on a particular assembly; this assembly is not always the current assembly. Additionally, all studies have not used the same assembly to do their analysis. In order to be able to compare data from different studies, it is necessary to obtain placements for all variants on the same set of assemblies. For most variants, we do this using the NCBI Remap Service using the following parameters:
- Minimum ratio of bases that must be remapped: 0.5
- Maximum ratio for difference between source length and target length: 4.0
- The 'Merge' function is turned on.
- Multiple locations can be returned.
We use relatively permissive parameters as many structural variants fall within regions of the genome that are likely to change from assembly to assembly. Additionally, NCBI Remap produces a coverage score that is calculated by taking the length of the feature in the target assembly and dividing it by the length of the feature in the source assembly. A score of 1 suggest the region is relatively unchanged between assemblies (note: single bases aren't assessed), a score of >1 suggests an insertion in the target assembly and a score of <1 suggests an deletion in the target assembly. We provide a qualitative assessment of the remapping status on our web pages:
- Perfect: coverage score equal to 1
- Good: coverage score within 5% of 1
- Pass: coverage deviates from 1 by greater than 5%
A relatively small number of variants submitted to dbVar are defined by sequence. These are typically insertion sequences that could not be placed on the assembly used in the analysis of the study. Such sequences are submitted to GenBank/EMBL/DDBJ and tracked in dbVar using the assigned accession.version. These sequences are aligned to newer assemblies using a process developed in-house called the NG Aligner (unpublished). Sequences that are aligned with >95% coverage and 98% identity are considered placed. Qualitative assessment of the remapping status is provided on our web page:
- Perfect:
- Good:
- Pass:
Assemblies in scope for each organism
We will always display placement data for the assembly used in the analysis of a study (the 'Submitted' placement). For most organisms, we will also attempt to find a placement for a variant on the 'current' assembly. When an assembly is updated we will support the 'current' and 'previous' assembly for up to a year. Human is an exception, we will support placements on NCBI36 and GRCh37 until GRCh38 is available.
FTP Site
All data in dbVar can be downloaded from our FTP site: ftp://ftp.ncbi.nlm.nih.gov/pub/dbVar/data/. Data is organized by assembly and by study. If you download by assembly, please be aware that some Variant Calls or Variant Regions may have been submitted on an assembly other than the one being downloaded; these variants will have been remapped to reflect their corresponding placements on the downloaded assembly. For example, the GRCh37 assembly download includes variants that were submitted on earlier assemblies all studies that were submitted on GRCh37, as well as all other variants, remapped to GRCh37. Remapped status is indicated in the download files.
If you choose to download instead by study, the data are available in three formats:
GVF:
- Variant regions and variant calls
- Variant regions and variant calls (compatible with UCSC Genome Browser)
- Variant regions only
- Variant regions only (compatible with UCSC Genome Browser)
TAB:
- Experiment
- Method
- Sample
- Sampleset
- Study
- Variant call
- Variant region
XML:
- Experiment
- Method
- Sample
- Sampleset
- Study
- Subject
- Variant Call
- Variant Region
dbVar Entrez search
A search bar is provided on the top of each dbVar page with a drop-down menu to search dbVar and other NCBI resources using Entrez (See Entrez Help for further details).
A dbVar search returns Studies, with links to the Study page, and Variants, with links to the Variant page. The filters on the right side allow the user to view All results, Study results or Variant results.
Limits
The dbVar Limits page allows the user to limit the search using check boxes rather than, or in addition to, using an Entrez query of search fields.
Advanced Search
The dbVar Advanced search page allows the user to build up an Entrez search query choosing the search fields and values from drop-down menus.
dbVar Search Fields
The following search fields are available to search dbVar:
Accession [ACC, ACCN]
Accession of any internal or external identifier. Versions removed from GENBANK accessions.
Accession Version [ACCV, ACCVERS]
Accession and version of GENBANK accessions used in variant sequence or support.
Allele Origin [ALOR, ORIGIN, ALLORGN]
Allele origin (controlled vocabulary), including Both=Germline+Somatic
All fields [ALL]
Default.
Assembly Accession [ASAC, ASSMACC, ASSMACCN, ASM_ACC]
Assembly accession of placement
Assembly Name [ASSM, ASSMBLY]
Assembly of placement
Assembly Organism [AORG, ASM_ORGN, ASM_ORG]
Assembly organism names (exploded)
Assembly Taxonomy ID [ATAX, ASM_TAXID, ASM_TAX_ID]
Assembly taxonomy ID
Author [AUTH]
All authors included in journal
Block Start [BLCK, BLOCK]
Start of a 100k block on chromosome containing the variant.
Chromosome [CH, CHRNUM, CHROM, CHROMOSOME]
Chromosome of placement
Chromosome Accession [CHRA, CHRACC, CHR_ACC, CHROM_ACCESSION]
Chromosome of placement, using accession.version
Chromosome End [CHRE, END, CHREND, CHR_STOP]
End of placement on chromosome
Chromosome Inner End [INRE, INNER_END, CHR_INNER_STOP]
Inner end of placement on chromosome
Chromosome Inner Start [INRS, INNER_START]
Inner start of placement on chromosome
Chromosome Outer End [OTRE, OUTER_END, CHR_OUTER_STOP]
Outer end of placement on chromosome
Chromosome Outer Start [OTRS, OUTER_START]
Outer start of placement on chromosome
Chromosome Start [CHRS, START, CHRSTART]
Start of placement on chromosome
Detection Method [DET, DETECTION]
Detection method
Discontinued Date [DDAT, DISCONTINUED, DISDATE]
dbVar discontinued date
Filter [FILTER]
Filter to return All records, just Study records or just Variant records.
Gene Full Name [GDSC, GENED, GENEFN, GENE_DESC, GENE_FULL]
Full name (description) of gene at same location as variant
Gene Name [GENE, GENE_NAME, SYM]
Name or alias of gene at same location as variant
Genome Projects ID [GPRJ, GENOMEPROJECT]
Unique identifier from Genome Projects
Genome Projects Name [PRNM, GPRJNAM, PROJECT]
Name from Genome Projects corresponding to Project_ID
Library Abbreviation [LIB, LIBRARY, LIBNAME]
Library name used in the Method
MeSH ID [MESH,MH]
Medical Subject Headings (MeSH) ID (exploded)
Method Platform [MPLT, METHPLAT, PLATFORM]
Method platform
Method Submission Name [MSUB]
Submission name of individual method, used when study contains multiple methods from different submitters, as does the curated dataset.
Method Type [METH, METHOD]
Method type (controlled vocabulary)
Method Type Category [MCAT, METHOD_CATEGORY]
Used for sorting and display. Methods are categorized as: probe, mapping, sequencing.
Method Type Weight [MWGT, METHOD_WEIGHT]
Used for sorting. BAC all Method_type values of study or variant are BAC aCGH, Non-BAC study or variant has at least 1 method_type that is other than BAC aCGH
MIM_id [MIM, OMIM]
MIM number.
Modification Date [MDAT, UDAT, UPDATE, UDATE, MODATE, UPDATEDATE]
dbVar Modification Date
Numeric Portion of EBI Study ID [ESTD, ESTNUM, ESTID]
Numeric portion of EBI Study ID (estd)
Numeric Portion of EBI Variant Call ID [ESSV, ESSVNUM, ESSVID]
Numeric portion of EBI Variant Call ID (essv)
Numeric Portion of EBI Variant Region ID [ESV, ESVNUM, ESVID]
Numeric portion of EBI Variant Region ID (esv)
Numeric Portion of NCBI Study ID [NST, NSTD, NSTNUM, NSTID]
Numeric portion of NCBI Study ID (nstd)
Numeric Portion of NCBI Variant Call ID [NSSV, NSSVNUM, NSSVID]
Numeric portion of NCBI Variant Call ID (nssv)
Numeric Portion of NCBI Variant Region ID [NSV, NSVNUM, NSVID]
Numeric portion of NCBI Variant Region ID (nsv)
Object Type [OT]
Object type in dbVar (STUDY, VARIANT)
Organism [ORG, ORGN]
Organism name (exploded)
Phenotype [PHEN, PHNO, PHENO, PTYPE]
Phenotype of sample/subject study or reference specimen
Placement Type [PTYP, PL_TYPE]
Placement type (controlled vocabulary)
PMID [PMID, PUBMED_ID]
Unique identifier from PubMed
Publication Date [PDAT, PUBDAT]
Journal Publication date
Sample [SMPL]
Sample/subject ID of study or reference specimen
Sample Count [SC, SCOUNT, SAMCOUNT]
Number of samples in study
Study Accession [ST, STACC, STUDY_ACCESSION]
Study dbVar ID (estd or nstd)
Study Description [STDE, STUDY_DESC]
Study description
Study Display Name [STDN, STUDY_DISP]
Study display name
Study ID [STDY, STUDY, STUDYID]
Study, batch or submission ID
Study Type [STYP, STYPE, STUDYTYPE]
Study type assigned by NCBI
Subject Phenotype status [SUPH, SUPSTA, SUB_PSTAT, SUB_PHENSTAT]
Boolean subject phenotype status: 0 not affected/null; 1 affected
Submitter Affiliation [LAB, CENTER]
Submitter's affiliation name
Submitter Name [SUB, SUBM]
Submitter first and last name
Submitter PDA Login [PDA, LOGIN]
Submitter login ID in NCBI PDA system
Submitter Variant ID [SVAR, SUBVAR, SVARID, SUBVARID]
Originally submitted variant identifier
Taxonomy ID [TXID, TAXID, TAXONOMY_ID, TAXONOMY]
Taxonomy ID
UID [UID]
UID
Unplaced Contig Accession [CTG, CONTIG, CTG_ACC, CTG_ACCESSION]
Contig of placement, when not on a chromosome, using accession.version
Validation Method [VAL, VALIDATION]
Validation method (controlled vocabulary)
Validation Result [VSTA, VSTAT, VALSTAT, VALIDATIONSTAT, VALIDATIONSTATUS]
Boolean validation status: null not validated, 0 validated with result 0; 1 validated with result 1
Validation Result Weight [VWGT, VALSTAT_WEIGHT]
0 not validated, 1 validated with result 0; 2 validated with result 1
Variant Call Accession [SSV, SSVACC, SSV_ACCESSION]
dbVar ID (essv or nssv) of Variant Call
Variant Call Count [SSVC, SSVCOUNT, SUPVARCOUNT, VC_COUNT]
Number of supporting variant calls in variant region
Variant Call Type [ALTP, ALLELE, VCTYPE, ALLTYPE, SSVTYPE]
Variant Call type (controlled vocabulary)
Variant Clinical Interpretation [CLIN, CLIN_SIG, CLIN_INT, CLINICAL_INTERPRETATION]
Clinical interpretation of a variant (controlled vocabulary)
Variant Description [DESC]
Variant description
Variant Region Accession [SV, SVACC, SV_ACCESSION]
dbVar ID (esv or nsv) of Variant Region
Variant Region Count [VC, VCOUNT, VARCOUNT]
Number of variant regions in study
Variant Region Type [VT, VTYPE, VARTYPE, VRTYPE, SVTYPE]
Variant region type (controlled vocabulary)
Variant Size [VLEN, VSIZE, VARSIZE, VARLEN, VARLENGTH]
Size of variant