| PubMed | All Databases | BLAST | OMIM | Books | Taxonomy | Structure |
| RefSeq Definitions |
RefSeq accession numbers can be distinguished from GenBank accessions by their distinct prefix format of 2 characters followed by an underscore character ('_'). For example, a RefSeq protein accession is NP_015325.
| Accession | Molecule | Method @ | Note | |
|---|---|---|---|---|
| AC_123456 | Genomic | Mixed | Alternate complete genomic molecule. This prefix is used for records that are provided to reflect an alternate assembly or annotation. Primarily used for viral, prokaryotic records. | |
| AP_123456 | Protein | Mixed | Protein products; alternate protein record. This prefix is used for records that are provided to reflect an alternate assembly or annotation. The AP_ prefix was originally designated for bacterial proteins but this usage was changed. | |
| NC_123456 | Genomic | Mixed | Complete genomic molecules including genomes, chromosomes, organelles, plasmids. | |
| NG_123456 | Genomic | Mixed | Incomplete genomic region; supplied to support the NCBI genome annotation pipeline. Represents either non-transcribed pseudogenes, or larger regions representing a gene cluster that is difficult to annotate via automatic methods. | |
| NM_123456 NM_123456789 |
mRNA | Mixed | Transcript products; mature messenger RNA (mRNA) transcripts. | |
| NP_123456 NP_123456789 |
Protein | Mixed | Protein products; primarily full-length precursor products but may include some partial proteins and mature peptide products. | |
| NR_123456 | RNA | Mixed | Non-coding transcripts including structural RNAs, transcribed pseudogenes, and others. | |
| NT_123456 | Genomic | Automated | Intermediate genomic assemblies of BAC and/or Whole Genome Shotgun sequence data. | |
| NW_123456 NW_123456789 |
Genomic | Automated | Intermediate genomic assemblies of BAC or Whole Genome Shotgun sequence data. | |
| NZ_ABCD12345678 | Genomic | Automated | A collection of whole genome shotgun sequence data for a project. Accessions are not tracked between releases. The first four characters following the underscore (e.g. 'ABCD') identifies a genome project. | |
| XM_123456 XM_123456789 |
mRNA | Automated | Transcript products; model mRNA provided by a genome annotation process; sequence corresponds to the genomic contig. | |
| XP_123456 XP_123456789 |
Protein | Automated | Protein products; model proteins provided by a genome annotation process; sequence corresponds to the genomic contig. | |
| XR_123456 | RNA | Automated | Transcript products; model non-coding transcripts provided by a genome annotation process; sequence corresponds to the genomic contig. | |
| YP_123456 YP_123456789 |
Protein | Mixed | Protein products; no corresponding transcript record provided. Primarily used for bacterial, viral, and mitochondrial records. | |
| ZP_12345678 | Protein | Automated | Protein products; annotated on NZ_ accessions (often via computational methods). | |
| NS_123456 | Genomic | Automated | Genomic records that represent an assembly which does not reflect the structure of a real biological molecule. The assembly may represent an unordered assembly of unplaced scaffolds, or it may represent an assembly of DNA sequences generated from a biological sample that may not represent a single organism. |
@ Method:
Mixed: indicates the process flow includes both automated processing and expert review for some of the records; curation analysis may be provided either by NCBI staff or collaborators.
Automated: indicates records that are not individually reviewed; updates are released in bulk for a genome.
|
|
The RefSeq COMMENT block indicates the Status of the record and the GenBank sequence data that was used to provide the record. In addition, the COMMENT may identify a collaboration that supplied the defining sequence information for the genome, gene, or protein. The level of curation may differ between different collaborating groups.
| STATUS | Definition |
|---|---|
| GENOME ANNOTATION | This identifies RefSeq records provided by the NCBI Genome Annotation process. These records are provided via automated processing and are not subject to individual review or revision between builds (see description of the assembly and annotation process). The mRNA records are identified based on alignments of other mRNAs to the genomic sequence and the proteins are conceptual translations of these mRNAs. These model transcripts and proteins may differ from pre-existing curated RefSeq (accession prefix NM, NR, NP) or GenBank records because they correspond to the genomic sequence. |
| INFERRED | Not curated. Inferred by genome sequence analysis with no direct same-species support for the product. Support for the record may include a combination of orthologous or paralogous protein homology and alignments of transcripts from related genes. A portion of the sequence may be defined by ab initio prediction. |
| MODEL | Not curated. The RefSeq record is predicted by a whole-genome computational genome annotation pipeline. The record may represent an ab initio prediction, or may have some level of transcript or protein homology support. |
| PREDICTED | Not curated. Automatically provided based on GenBank sequence data; limited or partial support for the transcript or protein. A portion of the transcript or protein may reflect an ab initio annotation prediction that was submitted to GenBank. |
| PROVISIONAL | Not curated. Automatically provided based on GenBank sequence data; there is support for the transcript and protein. This is the default status code applied to some genomes for which there is no clear information about the method used to define the sequence. |
| REVIEWED | Curated. The RefSeq record has been reviewed to provide the preferred sequence standard and to add additional functional descriptive information and feature annotation, as relevant. |
| VALIDATED | Curated. The RefSeq record has undergone an initial review to provide the preferred sequence standard. |
| WGS | Not curated. The RefSeq record represents a collection of whole genome shotgun (WGS) sequences. This status code is applied to genomic records. |
|
|
You can restrict your Entrez query to the RefSeq collection by using:
- Entrez Limits settings
- Entrez Property term restrictions
Using Entrez Limits:
You can use Entrez Limits settings to restrict your query to the RefSeq database. To use Entrez Limits you must first go to either the Nucleotide or Protein database; one way to do this is to query against all databases and follow links to the results in the desired database. [From the NCBI homepage, query against 'All Databases' or follow the link along the top bar to 'All Databases' and proceed from there.] Once you have navigated to Protein or Nucleotide results, note the Limits Tab located directly beneath the text area where a search term is entered.
| Description | ||
|---|---|---|
| this restricts the query to the RefSeq collection | ||
| this restricts the query to genomic RefSeq records | ||
| |
this restricts the query to mRNA RefSeq records |
The Entrez Limits page:
(click to open larger view of this image)
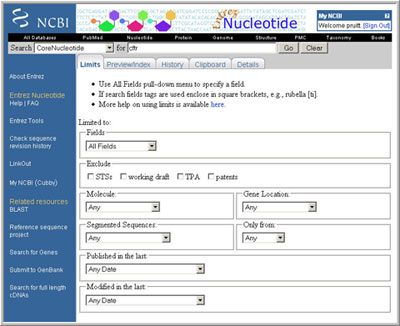
Refining a query using Entrez Properties restrictions:
More refined queries can be carried out to retrieve specific types of RefSeq records, such as those with a particular status (reviewed, etc.) or those from the genome annotation pipeline. The format for these queries is "term[prop]". You can review what terms are defined using the Entrez Preview/Index Tab located to the right of the Limits Tab.
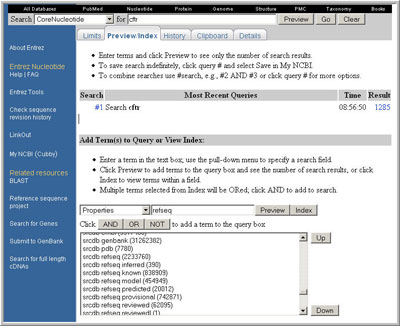
Find all of the property restriction terms that are defined for the RefSeq collection:
- navigate to the Preview/Index Tab
- select "Properties" from the menu
- enter 'refseq' or 'srcdb refseq' in the text field (without the quotes; 'srcdb' is an abbreviation of 'source database')
- click on the 'Index' button
- scroll through the resulting list to find the term(s) of interest (in this example, those beginning with 'srcdb refseq')
- add the restrictions to your query
To add a restriction to your query, select the term of interest (for example, 'srcdb refseq known') and click the appropriate Boolean button to configure the query as AND, OR, or NOT.
The Entrez Preview/Index page:
(click to open larger view of this image)
If you already know the property term, you can enter it directly into the search box as part of your query. The property terms defined for the RefSeq database and the accession prefixes that may be retrieved (per term) are listed below:
| Query Restriction | Accession Prefix Retrieved | Description | ||
|---|---|---|---|---|
| srcdb_refseq[prop] | NC_, AC_, NG_, NT_, NW_, NZ_, NM_, NR_, XM_, XR_, NP_, AP_, XP_, ZP_ | All NCBI RefSeq records
Try It: Nucleotide Protein |
||
| srcdb_refseq_reviewed[prop] | NC_, NT_, NW_, NG_, NM_, NR_, NP_, YP_ | reviewed records (curated)
Try It: Nucleotide Protein |
||
| srcdb_refseq_provisional[prop] | AC_, NC_, NT_, NW_, NG_, NM_, NP_, AP_, XM_, XP_ | provisional records (not curated)
Try It: Nucleotide Protein |
||
| srcdb_refseq_predicted[prop] | NG_, NM_, NR_, NP_, ZP_ | predicted records (not curated)
Try It: Nucleotide Protein |
||
| srcdb_refseq_validated[prop] | NC_, NG_, NM_, NR_, NP_, YP_ | validated records (curated)
Try It: Nucleotide Protein |
||
| srcdb_refseq_inferred[prop] | AC_, NG_, NM_, NP_ | inferred records (not curated); annotation inferred based on alignments from other genes or organisms
Try It: Nucleotide Protein |
||
| srcdb_refseq_known[prop] | NC_, NT_, NW_, NG_, NM_, NP_, AP_, YP_, ZP_ | reviewed, validated, provisional, predicted, inferred nucleotide or protein; excludes RefSeq records that are provided by the NCBI genome annotation pipeline (some NT_, NW_, and all XM_, XR_, XP_ accessions).
Try It: Nucleotide Protein |
||
| srcdb_refseq_model[prop] | NT_, NW_, XM_, XR_, XP_ | RefSeq records generated by the NCBI genome annotation pipeline (not curated); model records
Try It: Nucleotide Protein |
Examples:
The following examples illustrate how different information can be retrieved by querying against the NCBI Nucleotide or Protein databases. Note that queries can be restricted by using the Limits page, by adding the restriction using the Preview/Index Tab, or by typing a formatted query. To provide the links below, the Limit restrictions are translated into the equivalent property restriction in the URL; for example, the Limit of 'Molecule=Genomic' is converted in the URL into 'AND biomol_genomic[prop]'.
| Sample Query | Result |
|---|---|
| CoreNucleotide database, Limits Molecule=Genomic, Query= mitochondrial AND srcdb_refseq_reviewed[prop] | returns genomic mitochondrial RefSeq records that have a status of 'reviewed' |
| CoreNucleotide database, Limits Fields=Gene Name, Query=CFTR | returns GenBank and RefSeq nucleotide records that use the gene name of CFTR; Note a page tab is provided to review the RefSeq subset |
| CoreNucleotide database, Limits Molecule=Genomic, Query=human[organism] AND srcdb_refseq[prop] AND NC_000000:NC_999999[pacc] | by restricting to a specific accession series, this query returns the set of RefSeq human chromosome records corresponding to the reference genome |
| Protein database, Limits Fields=Gene Name, Query= CFTR AND srcdb_refseq[prop] | returns protein RefSeq records with a CDS feature annotated with gene=CFTR |
| Protein database, Query=srcdb_refseq[prop] AND "saccharomyces cerevisiae"[organism] | returns the set of S. cerevisiae RefSeq proteins |