Submitting data
GEO accepts many categories of high-throughput functional genomic data, including all array-based applications and some high-throughput sequencing data. This page summarizes deposit options and formats.
We aim to make data deposit procedures as straightforward as possible and will provide as much assistance as you require to get your data submitted to GEO. If you have problems or questions about the submission procedures, just e-mail us at geo@ncbi.nlm.nih.gov with a brief description of the type of data you are trying to submit, and one of our curators will quickly get back to you.
- Data types
- Submission format options
- Basic requirements for submissions
- Fast facts about submitting data
Submission format options Back to top
The table below summarizes GEO deposit formats. Deciding which method to use depends on the amount of data you have to submit, the format in which your data currently exist and what applications you are familiar with. Regardless of the deposit method you choose, your final GEO records will look the same and contain equivalent information.
| Option | Format | Key points | |
|---|---|---|---|
| GEOarchive | spreadsheets (e.g., Excel) |
Recommended method for most submissions. Quickly describe your study using Excel spreadsheet templates. Complete instructions » |
|
| SOFT | plain text |
Good option if your data and metadata are already in a database,
and you can generate and export data in SOFT plain text format. Complete instructions » |
|
| MINiML | XML |
Good option if your data and metadata are already in a database,
and you can generate and export data in MINiML XML format. Complete instructions » |
|
All deposit options described above can be used for any data type. However, the majority of GEO submitters use common commercial arrays (Affymetrix, Agilent, Illumina or Nimblegen) each of which has unique properties and file types. It is recommended that submitters who use the 4 common commercial arrays see these recommendations:
Basic requirements for array submissions Back to top
Your final GEO records will look something as follows:
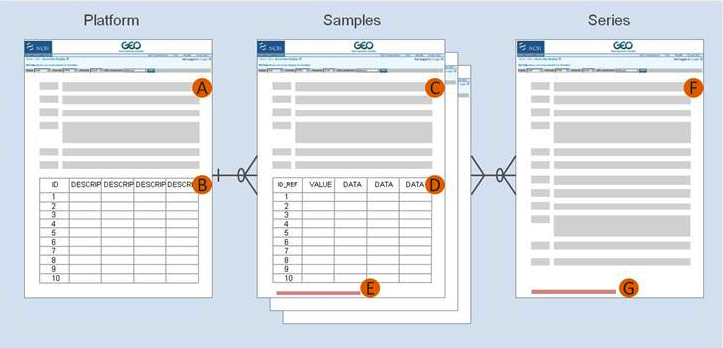
For almost all array data submissions, you will be asked to provide the following information:
| A | Text description of the array or sequencer |
|---|---|
| B | Text tab-delimited table of the array template |
| C | Text description of the biological sample and protocols to which it was subjected |
| D | Text tab-delimited table of processed hybridization result or sequence counts |
| E | Raw data file, or processed sequence data file |
| F | Text description of the overall experiment |
| G | Tar archive of raw data files, or processed sequence data files |
Fast facts Back to top
- All the deposit methods described above support MIAME-compliant data submissions. More...
- Regardless of the deposit method you choose, the final GEO records will look the same and contain equivalent information.
- Your GEO submissions may remain private until a manuscript describing the data is published. More...
- You can allow reviewers anonymous access to your private submissions. More...
- GEO accession numbers are normally approved within 5 business days after completion of submission. If you do not receive an e-mail from us within 5 business days of your submission, please first check your spam or junk e-mail folders because some systems recognize GEO e-mail correspondence as spam, then e-mail us at geo@ncbi.nlm.nih.gov to inquire about your submission. More...
- All the deposit methods described above support submission of many data types, including:
- For all studies involving human subjects, it is the submitter's responsibility to ensure that the data and files supplied to GEO protect participant privacy in accordance with all applicable laws, regulations and institutional policies. Make sure to remove any direct personal identifiers from your submission. These identifiers are listed in http://privacyruleandresearch.nih.gov/research_repositories.asp, footnote 1.
- You can update or edit your existing GEO records at any time using any of the formats described above, and regardless of the format in which they were originally uploaded. More...

