|
If you are familiar with GenBank sequence submission tools such as
Sequin, BankIt or tbl2asn, you can continue using these tools.
This step-by-step instruction is for GenBank submission of influenza virus sequences, using the Virus Wizard in the latest version of Sequin, a stand-alone
sequence submission tool.
1. Download and install Sequin on your computer if you do not have it or have an
older version than 12.21 (released on June 20, 2012).
2. Prepare a source table in tab-delimited text format containing sample information as shown below. For a template, you
can Download an Excel file, enter your own data in a spreadsheet program, and save the file in the
tab-delimited text format.
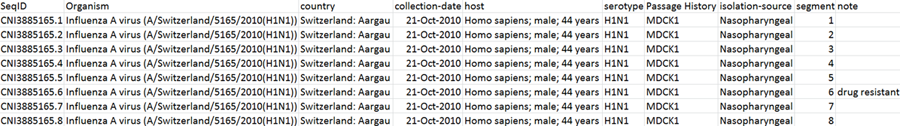
Instructions for making the table:
A. Headers of the table should be exactly the same as those in the example.
B. "SeqID" is required, and should be unique for each sequence and not longer than 25 characters. Avoid using symbols other
than letters, numbers or dashes.
C. Organism name should follow influenza naming convention (A/non-human host/location/isolate number/year in four digits(serotype)).
D. Use only GenBank standard country names in the "Country" field (e.g.
United Kingdom, not England). Sub-country location such as state and city can be included in this field.
E. Collection date should be in the format of DD-Mmm-YYYY (e.g. 03-Jul-2009).
F. Use scientific names (e.g. Homo sapiens) in the "Host" field whenever possible. Use lower cases for common names (e.g. duck), except for proper names (e.g. American wigeon). Host gender and age can be included in this field.
G. Additional source information can be included in the "note" column at the end of the table.
3. Prepare all nucleotide sequences in one FASTA
file, as shown in an example below. You can also download the
example file.
>CNI3885165.1
AGCGAAAGCAGGTCAAATATATTCAATATGGAGAGAATAAAAGAGTTAAGAGATCTGATGTCGCAGTCTCGCACTCGCGA
GATACTGACAAAAACCACTGTGGACCATATGGCAATAATAAAAAAATACACATCAGGAAGACAAGAGAAGAACCCCGCTC
TCAGAATGAAATGGATGATGGCAATGAAGTATCCGATTACAGCAGACAGGAGGATAATGGAGATGATTCCTGAAAGAAAT
...
>CNI3885165.2
AGCGAAAGCAGGCAAACCATTTGAATGGATATCAATCCGATTCTACTTTTCTTAAAAGTGCCAGCACAAAATGCTATAAG
CACTACATTCCCTTACACTGGAGATCCCCCATACAGTCATGGAACAGGGACTGGATACACCATGGACACGGTCAACAGAA
...
>CNI3885165.3
AGCAAAAGCAGGTACTGATTCAAAATGGAAGACTTTGTGCGACAATGCTTCAATCCAATGATCGTCGAGCTTGCGGAAAA
...
|
The definition line of each sequence should contain the SeqID used in the source table.
4. Validate your sequences using the NCBI Influenza Genome Annotation Tool, verify all "ERROR"s reported in the result page
and make corrections in the sequences if necessary. Once all sequences are error-free (except for the ones for PB1-F2 CDS, which are expected and
allowed), save the feature table of the annotation result on your computer.
5. Launch Sequin and click "Start New Submission". Enter required information in the "Submitting Authors" section.
6. Select "Viruses" for "Use a Submission Wizard".
7. Click "Import Nucleotide FASTA" and upload the FASTA sequence file prepared in Step 3.
8. Enter "Sequence Method" information.
9. Select "Submission Type" - this should usually be "Batch" for influenza sequences.
10. Select "Influenza virus" for "Virus Wizard Type of Virus".
11. Click "Import Source Table" and upload the source table file prepared in Step 2.
12. Select "cRNA" for "Virus Wizard Molecule Information".
13. Select "Multiple features per sequence" if "What do your sequences contain?" is asked.
14. Click "Upload Feature Table" and upload the feature table file prepared in Step 4.
15. Click "Open Record Viewer", make changes in the records if necessary and click "Done" to save the file.
16. Send the file as an attachment in an email to: gb-sub@ncbi.nlm.nih.gov.
After receiving these data, NCBI staff will start the GenBank submission process and communicate with submitters if there are any
issues with the submission.
Contact us if you have any questions.
| 


