By Dan Krotz
March 28, 2011
Role of American Recovery and Reinvestment Act (ARRA) Funding in This Discovery: The ARRA award was instrumental for the success of this project. The lead author of this study, Dr. Ben Montpetit, received salary support from ARRA, enabling him to focus on this project. Without the ARRA award, this project would not have been pursued at this time. In addition, Dr. Karsten Weis used ARRA funds to employ two additional postdoctoral fellows who were essential in completing this and other projects in his laboratory at the University of California, Berkeley (UC Berkeley).
Significance of This Project: Using a synchrotron that generates intense x-rays, researchers have produced a high-resolution map of the structure of an important molecular complex within cells. The molecular complex moves genetic information—in the form of messenger RNA—out of the nucleus and into the cytoplasm, where the RNA guides the production of proteins, the workhorses of biology. The researchers produced images of the structure of one particular protein, Dbp5, at several points in time as it remodeled messenger RNA for transport. They created a time-lapse movie of the RNA-exporting complex in action. The researchers also were able to identify the function of another molecule, IP6, that is involved in messenger RNA transport. IP6 stabilizes two other proteins long enough to kickstart Dbp5 into action. Each of these proteins serves important roles in the molecular complex.
By understanding the molecular interactions involved in these key steps of messenger RNA transport, the door opens to target treatments for diseases in which these molecules are involved.
Recovery Act Investment: "Posttranscriptional Regulation of Gene Expression in Eukaryotes"; Karsten Weis; University of California, Berkeley; 2009: $491,533 (1RC1GM091533-01); 2010: $499,985 (5RC1GM091533-02). Funded by the National Institute of General Medical Sciences.
Publications listing this Recovery Act Investment as providing grant support: Montpetit B, et al. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature, 2011;472(7342):238–242.
Onischenko E, Weis K. Nuclear pore complex-a coat specifically tailored for the nuclear envelope. Current Opinion in Cell Biology, 2011;23(3):293–301.
Munchel SE, et al. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Molecular Biology of the Cell, 2011;22(15):2787–2795.
Equipment Funded by Additional Recovery Act Grant:
Dr. Weis also received ARRA funding for a spinning disk confocal microscope for the UC Berkeley campus. This state-of-the-art microscope enables fast imaging of cells and tissues and is particularly useful for imaging cellular events in real-time and for extended time periods (i.e., hours to days). Such an instrument was previously not available to researchers on the UC Berkeley campus. The instrument has been installed in a shared microscopy facility in the Department of Molecular and Cell Biology at Berkeley. This microscope will not only be extremely beneficial to Dr. Weis’ research but it also will provide a tremendous resource for a large number of life science researchers on the UC Berkeley campus in general.
Recovery Act Investment: "Spinning Disk Confocal Microscope for the University of California, Berkeley"; Karsten Weis; University of California, Berkeley; 2010: $498,498 (1S10RR027696-01). Funded by the National Center for Research Resources.
A tiny motor tasked with one of nature’s biggest jobs is now better understood. The molecular machinery that helps export messenger RNA from a cell’s nucleus has been structurally mapped at the Advanced Light Source, a synchrotron located at the U.S. Department of Energy’s Lawrence Berkeley National Laboratory (Berkeley Lab).
Messenger RNA conveys genetic information from the nucleus to the cell’s cytoplasm, where it guides the synthesis of proteins — the workhorses of biology. A key protein complex that helps to ferry messenger RNA from the nucleus has been poorly understood until now, however.
“Our research describes how this protein complex works at the molecular level,” says Ben Montpetit, a postdoctoral researcher in Karsten Weis’ lab at the University of California, Berkeley. Their research, a collaboration with biochemists Nathan Thomsen and James Berger, also of the University of California, Berkeley, is described in a paper published March 27 in an advance online edition of the journal Nature. Berger is also a faculty scientist in Berkeley Lab’s Physical Biosciences Division.
The scientists studied a protein called Dbp5 that resides at the nuclear pore complex of fungi, plant, and animal cells. In these organisms, it reshapes messenger RNA as part of a chain of events required to send it from the nucleus.
But that’s just the tip of the iceberg. Dbp5 is among a class of enzymes called DEAD-box ATPase that perform vital RNA-remodeling functions throughout nature, from humans and oak trees to fungi and single-celled bacteria. Understanding how it works in the cells of one species will illuminate how it works in distantly related species.
“DEAD-box proteins are conserved throughout life, so learning how it works in this case sheds light on its function everywhere in nature,” says Montpetit.
The scientists conducted their research at beamline 8.3.1 of the Advanced Light Source, a national user facility that generates intense x-rays to probe the fundamental properties of substances. They used the synchrotron to resolve the structure of Dbp5 from yeast cells at key steps of the enzyme’s job, such as when it’s activated by another protein called Gle1 and when it binds with RNA. The structures were obtained at resolutions of between one and four angstroms (one angstrom is the diameter of a hydrogen atom).
The result is a time-lapse series of the protein’s choreographed bid to remodel messenger RNA, with its twists and turns revealed at the highest resolution yet.
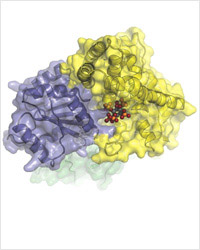
The closest look yet at the molecular machinery that helps transport messenger RNA from a cell’s nucleus. In this image, Dbp5 (blue-grey) and Gle1 (yellow) are glued together by IP6 (colored spheres). (Image courtesy of Karsten Weis’ and James Berger’s labs)
Among the team’s most intriguing discoveries is the role of a molecule that is known to be involved in messenger RNA transport, but whose function was a mystery. They found that the molecule, called inositol hexakisphosphate, or IP6, tethers Gle1 to Dbp5. This stabilizes the two proteins long enough for Gle1 to kickstart Dbp5 into action.
“IP6 acts like a molecular glue,” says Montpetit. “This is one of the first examples of an endogenous small molecule functioning to bring larger protein molecules together. With this knowledge, scientists can now consider how IP6 may be used to regulate mRNA export under various conditions, such as in response to stress.”
Their research could also advance scientists’ understanding of a rare but devastating family of diseases called lethal congenital contracture syndrome. The mutation that causes this disease is mapped to the genes that produce both Gle1 and IP6. Now, with Gle1’s role in messenger RNA transport further elucidated, the door opens for the development of therapies that target its function.
The protein complex in action. In this simulation, Gle1 (yellow) binds Dbp5 (green and blue-grey), causing Dbp5 to release RNA (orange). The movie begins with Dbp5 bound to RNA and then transitions to the Gle1-Dbp5 state, the step that jettisons RNA. The simulation then plays in reverse to mimic RNA and ATP dependent closing of Dbp5 and release of Gle1. (Movie courtesy of Karsten Weis’ and James Berger’s labs)
The research was funded by the National Institutes of Health’s National Institute of General Medical Sciences and the G. Harold and Leila Y. Mathers Foundation.
Lawrence Berkeley National Laboratory is a U.S. Department of Energy (DOE) national laboratory managed by the University of California for the DOE Office of Science. Berkeley Lab provides solutions to the world’s most urgent scientific challenges including sustainable energy, climate change, human health, and a better understanding of matter and force in the universe. It is a world leader in improving our lives through team science, advanced computing, and innovative technology. Visit our website.
Additional information:
- The research is described in a Nature article titled, “A conserved mechanism of DEAD-box ATPase activation by nucleoporins and IP6 in mRNA export.” It was published online March 27, 2011.
This article originally appeared on the University of California Lawrence Berkeley National Laboratory. Reposted with permission.