Staff Bibliography |
|
A Biomedical Information System for Combined Content-Based Retrieval of Spine X-ray Images and Associated Text Information
Sameer Antani, L. Rodney Long, George R. Thoma
U.S. National Library of Medicine
8600 Rockville Pike, Bethesda, MD 20894
Abstract
We present the status of ongoing work toward the development of a biomedical information system at the Lister Hill National Center for Biomedical Communications, a research and development division of the National Library of Medicine (NLM). For any class of biomedical images, the problems confronting the researcher in image indexing are (a) developing robust algorithms for localizing and identifying anatomy relevant for that image class and relevant to the indexing goals, (b) developing algorithms for labeling the segmented anatomy based on its pathology, (c) developing a suitable indexing and similarity matching method for visual data, and (d) associating the text information on the imaged person, indexed separately, for query and retrieval along with the visual information. We are in the process of building a content-based image retrieval system which supports hybrid image and text queries and includes a biomedical image database. The paper describes this prototype CBIR2 system and the algorithms used in it. Image shape-based retrieval is done by image example and user sketch of the vertebrae on the spine x-ray images from the National Health and Nutrition Examination Survey (NHANES) data.
Introduction
The general problem of developing algorithms for the automated or computer-assisted indexing of images by structural contents is a significant research challenge [1]. This is particularly so in the case of biomedical images, where the structures of interest are commonly irregular, and may be partially occluded. Examples are the images created by digitizing film x-rays of the human cervical and lumbar spines, digitized color slides of the uterine cervix, color endoscopy, endoscopic ultra-sonography, etc. Text data in the form of patient or survey data is commonly associated with medical images. Systems in use allow retrieval of image data through a text based query. Content-based image retrieval aims to allow researchers and medical practitioners access to the images directly by their content. We envisage that the development of a system that provides such access would have many applications in education, research, clinical trials, diagnosis, etc. For example, a medical school faculty member who is an expert in degenerative spine disease could query for examples of severe disc space narrowing for both sexes for the cervical spine; or a clinician could use it for searching for images similar to patient's present image pathology or injury.
The Lister Hill National Center for Biomedical Communications, a research and development division of the U.S. National Library of Medicine (NLM), maintains a digital archive of 17,000 cervical and lumbar spine images collected in the second National Health and Nutrition Examination Survey (NHANES II) conducted by the U.S. National Center for Health Statistics (NCHS). Along with the 10,000 cervical spine and 7,000 lumbar spine images, the NHANES II survey also included information on demographics, health questionnaire responses and physician's examination results. Over 2000 fields of information are available on each surveyed person, providing a large body of text information. Sub-sampled examples of the cervical spine x-ray is shown in Figure 1. Classification of the images for biomedical researchers, in particular the osteoarthritis research community, has been a long-standing goal of researchers at the NLM, collaborators at NCHS, and the U.S. National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), and capability to retrieve images based on geometric characteristics of the vertebral bodies is of interest to the vertebral morphometry community.

|
In this paper we describe the design of the prototype CBIR2 system that supports hybrid image and text queries. Image queries are posed by image-example or user-sketch. The CBIR2 system was developed after an initial feasibility study done through the development of the CBIR1 system [5]. This system was used to develop initial ideas on the algorithms, functionality, and interface characteristics of a CBIR system for digitized x-rays. The lessons learned from CBIR1 helped us establish clear objectives and define a superior architecture for the CBIR2 system.
The remainder of the paper is organized as follows. Section 2 outlines the research objectives. The design description, algorithms used and current status of our research is presented in Section 3. We summarize our work and project future work in Section 4.
Goals
The open research goals within the scope of the content-based biomedical image retrieval system include (i) content extraction, representation, feature classification, and similarity computation; and (ii) development of suitable query paradigms, indexing schema, and algorithms for efficient retrieval. For biomedical images the notion of image content, its representation, and similarity has an added dimension of relevance. It is very likely that the queries presented to the CBIR system are going to be specific to the pathology which may be subtle differences from what is considered to be normal. It is important that the extracted image features describing it are retained through the data-reducing feature representation process.
In general, our overall goal is to make a significant contribution to the image- and multimedia-rich digital library of the future by advancing CBIR techniques applied to the NHANES x-ray images, and eventually to other biomedical images. Specific objectives include:
- Conduct R&D in the steps needed for biomedical CBIR, viz., shape segmentation, feature extraction, feature vector organization and classification.
- Conduct R&D into query techniques suitable for biomedical images.
- Develop the algorithms needed to implement both indexing and retrieval.
- Design and develop a next-generation CBIR system (incorporating these algorithms).
- Develop a ground truth, evaluate the system, validate the results, and characterize the performance of the system.
- In the long term, extend CBIR techniques developed for the NHANES images to other biomedical images.
CBIR2 System Design and current Work
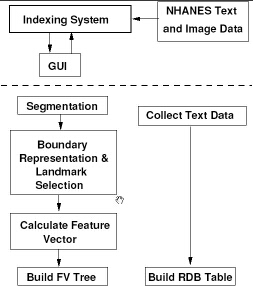
In this section we describe the design architecture of our current prototype system, CBIR2. The system designed in a modular fashion and is logically composed of two separate systems, viz., the indexing system and the retrieval system. The indexing system includes methods for automated image segmentation, image feature extraction, feature vector computation, feature organization, and text data organization. The retrieval system provides the interface and the methods for image and text retrieval. For this it includes methods for extracting features from example images, computing the feature vector, and determining similarity between features extracted from the query visual and those stored in the database. In addition, text retrieval via SQL and methods to combine the text and image queries are also included. The architectures for the indexing and retrieval systems are shown in Figures 2 and 4.
 |
The modular design of CBIR2 allows us to place simplified and possibly inefficient algorithms in place and yet have a completely developed testbed system while we wait on results from our research in each CBIR task.
Indexing system
The indexing process is currently semi-automated and done via a graphical interface. This interface allows indexing of two types of data. The text data is organized as fields in a relational database table from which data can be retrieved using the MySQL relational database manager. The indexing of the image data on the other hand is a more involved process. We describe the system modules here.
- Segmentation. The first step in indexing the images is the segmentation of the objects of interest, the vertebrae. The image quality in the spine x-ray images is fairly poor with ambiguous vertebral boundaries, making a reliable segmentation a challenging task. Current implementation contains human assisted boundary segmentation using Active Contour Segmentation (ACS). For this we have developed a tool shown in Figure 3. Our current research efforts into reliable shape segmentation include applying Active Shape Modeling (ASM) techniques [2] to extract the vertebra boundaries.
Figure 3: Active Contour Segmentation Tool: Main Window 
The ACS tool allows the user to place an initial template on the vertebra and apply the ACS algorithm. The user may enhance the image using histogram equalization before applying the method. The tool allows the user to create a template by marking points around the vertebra. The template can then be saved for future use. The position and size of the template can be controlled by rotation, translation and scaling prior to invoking the ACS algorithm. After the segmentation, the user can accept and save or discard the segmentation results. If the segmentation is accepted, the tool estimates the location of the next vertebra and places the template on it and the process is repeated. If at any point the segmentation is not acceptable, the user can perform manual segmentation on the vertebra. We have incorporated this feature in the tool to allow development of CBIR2 to proceed while we develop more reliable automated segmentation techniques. The tool generates segmentation output in a XML style file and is given a .cbr file extension. Here we store for each object, the template, and automated, and manual segmentation results. This way, entries can be modified as needed following future developments. The .cbr file records the information about an image, database source, view (e.g., lateral, sagittal, AP), and the human segmentor in the header structure. Additionally, the coordinate system origin is also specified. This is used by the objects within the image as a reference point. The image identifier is the same as that in the text database, allowing cross indexing across the image and text databases. The segmented objects are stored with a unique object identifier, anatomy identifiers, region of the anatomy, the segmented boundary points, the bounding box, the oriented bounding box, etc. The unique object identifier allows many versions of the segmented object to be retained. This has been designed so that a variety of database schema can be maintained. For example, the current best segmentation could be exposed for user CBIR searches, while research could proceed with other segmentation techniques. - Feature extraction and representation. The next step after shape segmentation in a CBIR system is representing the shape boundary information. The dense boundary points extracted as
 coordinates in the image space stored in the .cbr file need to be represented in a form suitable for archiving, indexing, and similarity matching. For this they are reduced to a small set of meaningful representative points by a shape representation algorithm. This coarse boundary and a binary image representation of the vertebra are used to find meaningful shape features that are invariant to translation, rotation, scaling and starting-point shift.
coordinates in the image space stored in the .cbr file need to be represented in a form suitable for archiving, indexing, and similarity matching. For this they are reduced to a small set of meaningful representative points by a shape representation algorithm. This coarse boundary and a binary image representation of the vertebra are used to find meaningful shape features that are invariant to translation, rotation, scaling and starting-point shift.
Most shape-based CBIR methods, to date, have been applied to art, trade-mark databases, fish images, silhouettes of tools, etc. From the literature, we observe that most shape representation methods use the global shape characteristics for indexing; i.e. the final shape representation is controlled by the distribution of all the boundary points in the image space. Such an approach may not be suitable for biomedical shapes due to (i) high similarity across anatomical shapes, such as vertebrae (ii) loss of subtle differences in boundary representation which could be indicative of certain pathology, and (iii) in adequacy of shape representation methods for supporting region localized queries.
The methods selected are briefly described below and are all currently implemented in CBIR2. We are in the process of developing other more suitable shape representation approaches. - Global Shape Properties and Invariant Moments. Global shape properties and moments are features intrinsic to a shape. The properties used are major and minor axis length and angle, compactness, roughness, and elongation. In order to compute these we converted the shape contour to a binary image and gave the same weight for each pixel inside the shape contour boundary. In addition, we also used first and second order 2D invariant moments defined by Hu [4].
- Scale Space Filtering. Scale space filtering reformats the shape boundary points to represent the shape at different levels of detail. It is said to follow human perception of shapes [3]. It provides more detail at scale higher level and progressively reduces the detail level until the shape becomes an oval shape. While capable of shape matching, a problem with this method is that the shape shrinks as it progresses from high detail level toward low detail level making comparison scale sensitive.
- Polygon Approximation. Polygon approximation or curve evolution is a process that eliminates insignificant shape features and reduce the number of data points. The resultant representation is one that uniquely describes the shape. The approximated curve was then converted to tangent space for similarity measurement [6].
- Fourier Descriptors. The position of a point on a closed contour is a periodic function. Thus, the Fourier series may be used to approximate the contour. The resolution of the contour is approximation is determined by the number of terms in the Fourier series. Since simple operations such as scaling and translation are related to simple operations of the boundary's Fourier descriptors, they are attractive for use with boundary matching [12]. Rotation invariance computation, however, requires the bend angle function to be computed.
- Feature organization. A feature vector is then created from various computed features and organized into a data structure for efficient retrieval. The development of a feature organization strategy is strongly correlated with the feature vector used, the query types supported, and the image semantics. We are at a stage where we have some of the these requirements identified. While research is proceeding toward determining an effective feature vector organization strategy, we are currently using a flat structure and linear search for retrieval. Having an inefficient, but working system enables us to improve on various modules as the research evolves.
- Feature classification. Our work toward the indexing of spine images for features of interest in the osteoarthritis and vertebral morphometry research communities requires the segmentation of the images into vertebral structures with sufficient accuracy to distinguish pathology on the basis of shape, labeling of the segmented structures by proper anatomical name, and classification of the segmented, labeled structures into groups corresponding to high level semantic features of interest, using training data provided by biomedical experts. We have adopted a hierarchical approach to such indexing that consists of high-level region classification, spine region localization, vertebra localization and identification, vertebral segmentation, and classification of the vertebrae by presence/absence of the biomedical features above [7,8,9]. Classification of the vertebra for biomedical features is being collaboratively done with Stanley [10,11]. However for shape based retrieval, labeling, classification, and extraction of features of interest, the vertebra boundaries need to be segmented from the images.
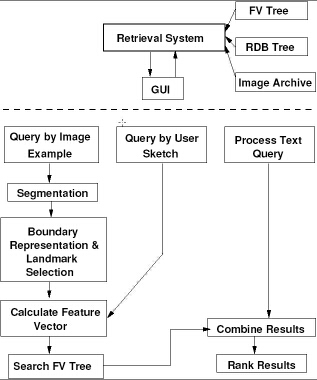 |
Retrieval system
As mentioned before, CBIR2 is logically organized as indexing and retrieval systems. However, in reality, the interface presented to the user is the ``retrieval'' system. The indexing process can be conducted through the same interface. The system is implemented in MATLAB. The basic types of queries supported are to the text data, image data and combined queries to both. The retrieval of the text data is supported through Open-Database Connectivity (ODBC) protocol to retrieve results using the MySQL DBMS. The queries to the image data can be specified in using an example image to retrieve images that are visually similar or by drawing a sketch of the indexed feature, in this case the vertebra boundary. The system presents the user with a GUI for creating queries and supports text, image example, and image sketch queries, and queries that combine text and image example or image sketch.
 |
 |
Figures 5 and 6 show the initial screen and the options screen for generating the basic query. A feature in CBIR2 allows users to save and recall their queries. The retrieval paths for image-example based queries and sketch-based queries are the same except for the feature extraction phase necessary for the former.
 |
The same feature extraction phase as in the indexing process is applied to the example image. The user is presented with the ACS tool for segmenting the image. The extracted image features in the query are then matched by a shape similarity algorithm to determine the similarity distance between the query and the database shape. The greater the distance between two feature vectors the greater is the dissimilarity. The system allows users to specify an image for an image-example based query as shown in Figure 7. For an sketch-based query, the users may choose to either use one of the provided templates, or use their own template and modify it or draw an outline from scratch. The user can save the template for a future use. The dialog boxes for these are shown in Figures 8 and 9.
 |
 |
It is also possible to narrow the query to the segmentation done by a particular algorithm. This is because the .cbr file allows the user to store several segmentations, including manual segmentations. Additionally, with several shape representation methods included, the user can also query on a specific representation. This feature is extremely useful in conducting evaluations for segmentation and representation algorithms. The system also reports a profile of the execution time taken for making a database query, computing the similarity, etc. This will be useful in evaluating overall system performance and advantages gained through better feature organization. The results from the query are presented as shown in Figure 10.
 |
Conclusions and Future work
In this paper we have described the work in progress toward building a complete biomedical information system. The system has a multimedia component and a text database component. This is in line with our eventual goal of developing a system that not only performs conventional text queries, provides content-based image retrieval queries, but also provides the user with meaningful information from the image classifications. The CBIR2 prototype described above provides a testbed for this.
The development of CBIR2 allows us to investigate various tasks at hand towards achieving the goals identified earlier. Automated shape segmentation and the issue of validation of the segmented boundary are yet to be addressed. The shape representation and similarity methods are limited in their abilities; i.e., while they allow matching of the entire vertebra shape, it is not possible to pose queries on local features of importance such as anterior osteophytes. Additionally, we are still in the process of identifying a suitable feature organization structure. Finally, development of a ground truth for evaluation and system performance characterization is planned.
Bibliography
1. S. Antani, R. Kasturi, and R. Jain.
A survey on the use of pattern recognition methods for abstraction,
indexing and retrieval of images and video.
Pattern Recognition, 35(4):945-965, 2002.
2. T. F. Cootes and C. J. Taylor.
Statistical models of appearence for computer vision.
Technical report, University of Machester, Wolfson Image Analysis
Unit, Imaging Science and Biomedical Engineering, University of Manchester,
Manchester, M12 9PT, U.K., February 2001.
3. A. Del Bimbo and P. Pala.
Shape indexing by multi-scale representation.
Image and Vision Computing, 17(3-4):245-261, 1999.
4. M. K. Hu.
Visual pattern recognition by moment invariants.
IRE Transactions on Information Theory, 8:179-187, 1962.
5. D. M. Krainak, L. R. Long, and G. R. Thoma.
A method of content-based retrieval for a spinal x-ray image
database.
Proceedings of IS&T/SPIE Medical Imaging 2002: PACS and
Integrated Medical Systems, 4685:108-116, February 2002.
6. L. J. Latecki and R. Lakämper.
Shape description and search for similar objects in image databases.
In R. C. Veltkamp, H. Burkhardt, and H. P. Kriegel, editors, State-of-the-Art in Content-Based Image and Video Retrieval, volume 22 of
Computational Imaging and Vision, pages 69-96. Kluwer Academic Publishers, 2001.
7. L. Long, D. Krainak, and G. Thoma.
Identifying image structures for content-based retrieval of digitized
spine x-rays.
In Proceedings of IS&T/SPIE Medical Imaging 2002: Image
Processing, Vol. SPIE 4684, pages 1204-1214, San Diego, CA, February 2002.
8. L. Long and G. Thoma.
Computer-assisted retrieval of biomedical image features from spine
x-rays: Progress and prospects.
In Proceeding of the ![]() IEEE Symposium on Computer-Based Medical Systems (CBMS 2001), pages 46-50, Bethesda, MD, July 2001.
IEEE Symposium on Computer-Based Medical Systems (CBMS 2001), pages 46-50, Bethesda, MD, July 2001.
9. L. Long and G. Thoma.
Landmarking and feature localization in spine x-rays.
Journal of Electronic Imaging, 10(4):939-956, 2001.
10. R. J. Stanley, M. Cherukuri, L. R. Long, and G. R. Thoma.
The application of shape features to cervical spine vertebra image
analysis.
Submitted to IEEE Transactions on Medical Imaging, 2002.
11. R. J. Stanley and L. R. Long.
A radius of curvature-based approach to cervical spine vertebra image
analysis.
In ![]() Annual Rocky Mountain Bioengineering Symposium, volume 37, pages 385-390, 2001.
Annual Rocky Mountain Bioengineering Symposium, volume 37, pages 385-390, 2001.
12. C. Zahn and R. Roskie.
Fourier descriptors for plane closed curves.
IEEE Computer, C-21(3):269-281, 1972.








