Within the National Cancer Institute (NCI), there is new potential for an increasingly beneficial collaboration between the Cancer Imaging Program (CIP) in the Division of Cancer Treatment and Diagnosis (DCTD) and the Center for Biomedical Informatics and Information Technology (CBIIT) in the Office of the Director. The Institute’s recent launch of the National Cancer Informatics Program (NCIP), which is being administered through CBIIT, provides this opportunity since one of the new program’s goals is to maintain and extend successful partnerships that had been forged under the auspices of the cancer Biomedical Informatics Grid® (caBIG®) program.
Beginning in 2005, CIP provided funding and expertise to CBIIT to help establish an In Vivo Imaging Workspace as part of the caBIG® program. Both CIP and those of us who formed the core of the Imaging Workspace envisioned building a wide-ranging, coherent portfolio of imaging research. This portfolio would catalyze the development and validation of open-source applications and algorithms to acquire, store, annotate, analyze, integrate, interpret, and share quantitative imaging data for the first time in the history of cancer imaging. The Imaging Workspace developed a suite of software applications that have supported The Cancer Imaging Archive (TCIA), which is now a major resource that CIP provides to the extramural research community. TCIA uses the National Biomedical Imaging Archive (NBIA) software as the core of its service and hosts more than 40 highly targeted collections of imaging phantoms and clinical images for research purposes, more than 20 of which are publicly accessible. These are now being accessed by more than 1,400 registered users.
These partnerships and others that coalesced around CIP and CBIIT/caBIG® will continue evolving within the context of the NCIP. We also believe that this collaboration provides a foundation that suggests how CBIIT/NCIP can work to establish viable partnerships with other intramural programs or projects across the NCI.
CIP and the Imaging Workspace share the goal of developing and disseminating imaging informatics solutions across the cancer research continuum, from basic and preclinical studies (including small-animal imaging) through translational and clinical research. This development continuum also requires standardizing and validating research algorithms and technologies for analyzing enormous amounts of imaging data and metadata in ways needed to satisfy the regulatory requirements set by the U.S. Food and Drug Administration (FDA).
One of the most recent CIP initiatives CBIIT has begun working to support is the Quantitative Imaging Network (QIN). To date, the network comprises fifteen academic research teams sharing the goal of developing an informatics architecture that will allow them to share imaging data and metadata produced by current commercial imaging platforms, integrate these data types with clinical data, and put the data to secondary use (primarily validation and qualification of imaging biomarkers of response to therapy and prognosis that could be used in clinical practice). QIN is focused on developing more standardized methods for image data collection and analysis across many different commercial imaging platforms. This will permit quantitative correlation with other omics data and provide the new NCI National Clinical Trial Network with advanced tools to support precision medicine. The CBIIT Imaging Workspace, CIP, and QIN share a strong commitment to achieving informatics and data interoperability standards, which are required for clinical decision-support systems.
The NCIP portfolio of open-source imaging informatics capabilities includes NBIA, Annotation and Image Markup (AIM), In Vivo Imaging Middleware (IVIM), the eXtensible Imaging Platform (XIP), and the Algorithm Validation Tool (AVT), which are collectively known as the Imaging Suite. QIN not only uses the NBIA-enabled TCIA to share its data within the network, but is also exploring the use of AIM to manage its metadata. Individual QIN sites use a variety of other open-source or proprietary imaging informatics applications, including for example 3D Slicer, which is being extended to store markup in the AIM format. A major advantage that the AIM application holds for the QIN is that it allows for a standard structure for storing imaging annotations.
From the point of view of the CBIIT Imaging Workspace, the deployment of our applications through various CIP projects has been of enormous benefit. It has allowed us to receive feedback on how effectively these tools meet scientific requirements—this is vital information we use to guide the further development of software that can better answer specific research needs. However, as QIN research expands to other imaging modalities and advanced tools, much work still needs to be completed to make quantitative imaging and its implementation in adaptive therapy trials a clinical reality.
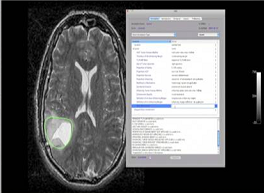
Another important CIP project supported by CBIIT and the Imaging Workspace involves the collection of imaging data from TCGA cases in a central repository (TCIA), and the facilitation of teams of investigators who are deriving computable data from the images and looking for correlations with the genomic and clinical data available through the TCGA data portal. Researchers involved in this effort are using distributed AIM-enabled workstations developed by the CBIIT Imaging Workspace to generate imaging characterization scores. The TCGA-GBM (glioblastoma multiforme) collection includes data on more than 240 de-identified GBM subjects. The project’s goal is to correlate morphologic imaging features with various gene-expression pathways relevant to disease progression and thereby identify candidate biomarkers. Their efforts have resulted in more than two dozen abstracts and several publications during the past two years. This unique resource and the CIP/CBIIT collaboration could play an important role in the development of more advanced, operator-independent image-processing methods for large-scale analysis of images and genomic data.
CBIIT’s contribution to developing tools and methods combined with CIP’s 14-year history of working with the imaging research community will continue to produce important advances in imaging informatics and research in such areas as the development of clinical imaging decision-support applications, a CIP priority. Our collaboration with CIP will continue to ensure that the development of tools and methods remains driven by the research needs of the community. We are now forming a new NCIP Imaging Working Group that will help ensure that the informatics solutions we help develop directly facilitate research across the basic, translational, and clinical continuum.
NOTE: The images accompanying this blog posting were taken from a Powerpoint presentation delivered by E. Helton, E. Siegel, and P Mulhern at the annual meeting of the American Association for Cancer Research.
Authors
Edward Helton, Ph.D., Associate Director for Clinical Imaging, NCI Center for Biomedical Informatics and Information Technology
Kay Fleming, Ph.D., Scientific Writer, NCI Center for Biomedical Informatics and Information Technology
Suggestions for Further Reading
Hwang SN, Clifford R, Huang E, Hammoud D, Jilwan M, Raghavan P, Wintermark M, Gutman DA, Moreno C, Cooper L, Freymann J, Kirby J, Krishnan A, Dehkharghani S, Jaffe C, Saltz JH, Flanders A, Brat DJ, Holder CA, Relationship between MR Imaging Features, Gene Expression Subtype, and Histopathologic Features of Glioblastomas. Poster presented at the Radiological Society of North America (RSNA) 2011 Annual Meeting: November 27–December 2, 2011, Chicago, IL.
Jain R, Poisson L, Naranj J, Scarpace, Rempel S, Mikkelsen T, Correlation of perfusion parameters with genes related to angiogenesis regulation in glioblastoma: a feasibility study, American Journal of Neuroradiology, 2012;33(7):1343-8.
Levy MA, Freymann JB, Kirby JS, Fedorov A, Fennessy FM, Eschrich SA, Berglund AE, Fenstermacher DA, Tan Y, Guo X, Casavant TL, Brown BJ, Braun TA, Dekker A, Roelofs E, Mountz JM, Boada F, Laymon C, Oborski M, Rubin DL, Informatics methods to enable sharing of quantitative imaging research data, Magnetic Resonance Imaging, 2012:in press.
The Vasari Research Project: https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Project
The Quantitative Imaging Network (QIN): http://grants.nih.gov/grants/guide/pa-files/PAR-08-225.html






The collaboration of these two institutes will be of great help for the continuous research on how to cure cancer. This can be also be beneficial in making a more accurate diagnosis for medical practitioners and help them make more advance researches which might be someday will lead on the discovery for the cure for cancers.
Sir, recently i heard about a cancer detection test. That is fecal occult blood tests (FOBT), actually how this test conduct. and bu doing this can we determine all type of cancers? . Waiting for your reply
sincerely
Navas Azeez
The primary public health and education website of the National Cancer Institute does contain some basic information on this test. Please visit: http://www.cancer.gov/cancertopics/pdq/screening/colorectal/patient/page3#Keypoint7. There are additional links for more information there as well.
Best,
Lisa M. Cole
Director of Communications
NCI CBIIT