Derivation of neural crest cells from human pluripotent stem cells.
Title |
Derivation of neural crest cells from human pluripotent stem cells. |
Date Submitted |
April 20, 2012 |
Submitted by |
Kim, Hyesoo (hkim179@jhmi.edu) |
Adapted from |
Lee et al, Nature Protocol, 2010 |
Contributor |
Lee, Gabsang. Chambers, Stuart. Tomishima, Mark. Studer, Lorenz. |
Affiliation |
Sloan-Kettering Institute |
Introduction:
Neural crest (NC) cells are a transient, multipotent, migratory cell population unique to vertebrates that emerge at the interface between neural and non-neural ectoderm, and migrate extensively to form a variety of NC derivatives, including peripheral neurons, glia, melanocytes, endocrine cells, craniofacial cartilage and bone, smooth muscle and enteric neurons.
Access to reliable protocols for the differentiation of human ES cells (hESCs ) and human induced pluripotent stem cells (hiPSCs) into NC lineages provides a model system to study normal development and also to interrogate the impact of genetic disease during NC development, such as Hirschsprung's disease, DiGeorge syndrome, Waardenburg syndrome, Charcot-Marie-tooth disease, familial dysautonomia, CIPA (congenital insensitivity to pain with anhidrosis) and pediatric cancers, such as neuroblastoma. In addition, enriched populations of defined NC cell types may be used as a source for application in regenerative medicine such as the repair of peripheral neuropathy or cranial skeletal defects.
These protocols for the induction of NC cells derived from hESCs and for the isolation and expansion/differentiation of hESC-derived NC cells were published on Nature Protocol by Lee et al. Other than hESCs, the protocols are also applicable to hiPSC lines derived from somatic cells such as normal fibroblasts, as well as fibroblasts derived from patients with NC-related genetic disorders.
Flowchart:
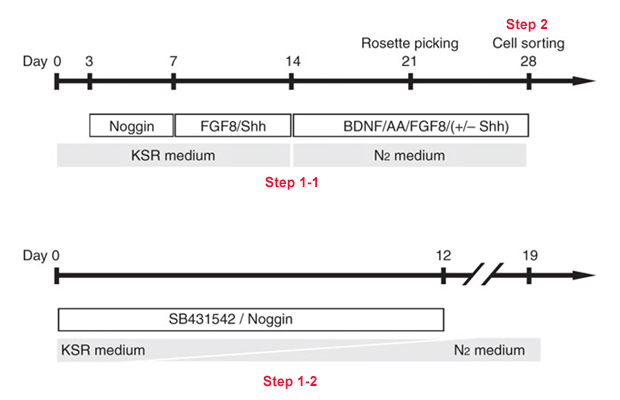
Protocol
1-1. Induction of NC cells using MS5 co-culture
- When human pluripotent stem cells (hPSCs) are confluent, discard hESC medium from tissue culture dish (60 mm tissue culture dish) and add 2mg/ml of dispase (2ml).
- Incubate at 37 °C for 15–20 min. and check under the microscope. When the edges of the hPSC colony start to detach (“rolling up”), harvest the colonies in fresh hESC medium. Under these conditions, most of colonies should detach, whereas PMEFs should remain attached.
- Centrifuge the harvested hPSCs for 5 min at 160g at room temperature and aspirate the supernatant.
- Repeat Step 3).
Critical step Repeated washing is necessary to dilute out dispase because dispase is not inactivated by serum. Incomplete washing out of dispase prevents hESC colonies from attachment and leads to high levels of cell death. - Pipette and triturate the hPSC pellet with 1 ml of hESC medium using P1000 micropipette.
Critical step The number of repeats and the intensity of pipetting determine the size of hPSC colonies. Colonies that remain too large tend to spontaneously differentiate, whereas hPSCs triturated too harshly tend to die on replating. - For subculture (maintenance of undifferentiated hPSCs), plate hPSCs onto fresh PMEF plates at a ratio of 1:5 to 1:8. Incubate at 5% CO2 and 37 °C. It is important to assure that hPSC colonies are dispersed evenly within the plate by carefully shaking dishes.
- For differentiation (day 0) (neural induction of hPSCs), plate small fraction of hPSCs (~ 5 × 103 to 30 × 103 cells per 60 mm dish) onto mitotically inactivated MS-5 plate (gelatin-coated 60 mm dish, ~1.2 million MS5 cells per dish, plate them a day before) (For Gelatin-coated dishes, coat culture dishes or flasks with sterile gelatin solution (0.1% (wt./vol) in water) for about 5 min at room temperature. Aspirate the gelatin solution before plating the cells)(P0).
Critical step The number of hPSCs in a MS-5 dish (60 mm dish) corresponds to ~2% of the undifferentiated hPSCs harvested from a 60 mm dish before passage. - From days 3 to 7, change KSR medium supplemented with 500 ng/ml of Noggin* every 2–3 d.
*500 ng/ml of Noggin can be replace with LDN193189 (100nM). - At day 7 and 12, change KSR medium supplemented with Shh (200 ng/ml) and FGF8 (100 ng/ml).
- From days 12 to 24, change Neurobasal medium supplemented with AA (0.2 mM), Shh (200 ng/ml), FGF8 (100 ng/ml) and BDNF (20 ng/ml) every 2–3 d. The combination of cytokines/growth factors added at late P0 (passage 0) stage (after Step 9)) and P1 (passage 1) stage (Step 11)) can be fine-tuned to increase NC yield. Cytokines and compounds controlling efficiency of NC induction in hESCs that have been described previously include Wnt3A (40 ng/ml), BMP4 (50 ng/ml), FGF8 (100 ng/ml), RA (0.5 μM), Shh (200 ng/ml), SU5402 (10 nM), Dickkopf-1 (100 ng/ml) and Noggin (500 ng/ml).
- At ~12–20 d of differentiation, numerous neural rosette structures emerge from the differentiating hPSCs (Fig. 1). At the latest by day 28 of differentiation, harvest rosette structures mechanically by aspirating colonies using a 1 ml syringe with a fine (27 G) needle (P0) and gently replate colonies on 15 μg/ml PO/1 μg/ml Lam/10 μg/ml FN-coated culture dishes in Neurobasal medium supplemented with AA (0.2 mM), Shh (200 ng/ml), FGF8 (100 ng/ml) and BDNF (20 ng/ml) (P1). Change medium every 2–3 d.
Critical step For preparing PO/Lam/FN-coated plate, coat 15 μg/ml PO in PBS in a 60 mm dish and keep it aside overnight. Next day, wash the PO-coated dish with PBS and coat 1 μg/ml laminin and10 μg/ml fibronectin in PBS for more than 2 h (usually overnight). Just before plating rosette structure, dry the PO/Lam/FN-coated dish.
Figure 1 Procedures for isolating hESC-derived neural crest cells with MS5 coculture. Undifferentiated (a) hESCs are induced to neural lineages (b–f, neural rosette) in the presence of appropriate morphogens/cytokines. (Inset in f) NC markers (AP2 and p75) are typically expressed at the periphery of neural rosette structure. Expression of the CNS marker Pax6 is observed in neural rosettes. Scale bars=100 μm.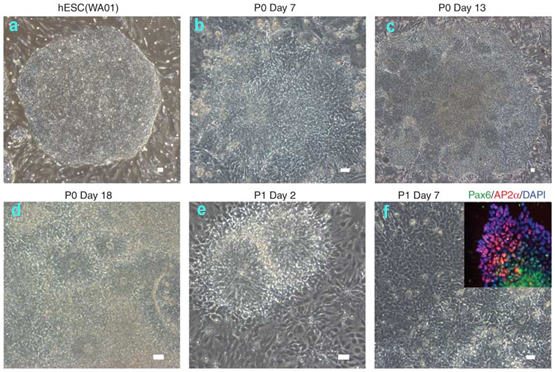
1-2. NC differentiation from hPSC using defined NSB culture system
- When hPSCs are confluent, aspirate the media and add a minimal amount of accutase solution needed to cover the surface of the dish.
- Incubate the dish at 37 °C for 15–30 min and check under the microscope. When all cells are rendered to single cells, harvest the colonies in fresh hESC medium.
- Avoiding air bubbles, triturate the cells in the dish using a Pasteur pipette with additional hESC media until the cells are in single cell suspension and filter using a 40 μm cell strainer to remove debris and cell clumps.
- Wash and centrifuge cells for 5 min at 160g at room temperature in hESC media to remove any remaining accutase solution.
- Resuspend the cells in hESC media containing ROCK inhibitor and plate on a gelatin-coated dish at a density of <200,000 cells per cm2.
- Incubate the dish for 30 min at 37 °C in an incubator in the presence of hESC media containing ROCK inhibitor. Prepare matrigel-coated dishes while the cells are incubating.
Critical step For coating Matrigel, dilute a 1 ml frozen aliquot of matrigel in 19 ml of DMEM: F12. A 45-μm cell strainer can be used to remove any insoluble clumps. Coat dishes with the diluted matrigel solution and let them stand for 1 h. Aspirate the matrigel solution and rinse dishes once with DMEM: F12 before plating the cells. - Collect the nonadherent cells, wash the dish with hESC media containing ROCK inhibitor and centrifuge the cells.
- Resuspend the cells in MEF conditioned media (CM) containing ROCK inhibitor.
Critical step For MEF conditioned media, thaw, rinse and plate two vials of MEFs (around 15 million cells) on a gelatin-coated T225 flask in DMEM with 10% FBS. The next day, aspirate the media, wash the dishes with hESC media to remove residual serum and plate 100 ml of hESC media. After 24 h, collect the MEF conditioned media (CM), and replace with new hESC media. This can be repeated daily for no more than 10 d. The media may be stored at 4 °C for less than 1 month. Before using, CM can be sterilized by 0.22-μm filtration and supplemented with 10 ng/ml of FGF2. - Determine the cell concentration using a hemacytometer and add CM containing ROCK inhibitor to the appropriate cell concentration.
- Plate cells on matrigel-coated dishes at 10,000 cells per cm2.
- Grow cells in CM containing 10 ng/mlFGF2, feed daily. In addition, for the first 2 d, CM should contain ROCK inhibitor.
- When the cells are 50–70% confluent, differentiation should be initiated (Fig. 2). To initiate differentiation, replace the media with KSR medium containing 10 μM SB431542 and 500 ng/ml of Noggin. Replace the media and slowly switch from KSR to Neurobasal media on days 2, 3, 5, 7, 9 and 11, and as detailed below.
Critical step Cell density at the time of initiation of differentiation determines the relative amounts of CNS versus NC cells produced, with lower confluency biasing toward increased numbers of NC cells and high confluency biasing toward CNS lineage. However, at very low confluency (<50% confluent), cell viability diminishes. Initial seeding densities ('plating efficiency') can vary between pluripotent cell lines and should be empirically determined to reach ideal starting conditions.

Figure 2: Target hESC/hiPSC density for NSB induction toward NC cells. The initial density of pluripotent cells will impact the amount of NC cells formed. Shown is the ideal density for starting the NSB induction to achieve NC cell formation. Scale bar=100 μm. - On day 2 of differentiation, aspirate the KSR using a sterile glass pipette and add KSR containing 10 μM SB431542 and 500 ng/ml of Noggin.
- On day 3 of differentiation, aspirate the KSR using a sterile glass pipette and add KSR containing 10 μM SB431542 and 500 ng/ml of Noggin.
- On day 5 of differentiation, aspirate the KSR using a sterile glass pipette and add media (75% KSR, 25% Neurobasal) containing 10 μM SB431542 and 500 ng/ml of Noggin.
- On day 7 of differentiation, aspirate the KSR using a sterile glass pipette and add media (50% KSR, 50% Neurobasal) containing 10 μM SB431542 and 500 ng/ml of Noggin.
- On day 9 of differentiation, aspirate the KSR using a sterile glass pipette and add media (25% KSR, 75% Neurobasal) containing 10 μM SB431542 and 500 ng/ml of Noggin.
- On day 11 of differentiation, aspirate the KSR using a sterile glass pipette and add NEUROBASAL media containing 10 μM SB431542 and 500 ng/ml of Noggin.
- NC cells can be observed by day 11 of differentiation based on AP2a, p75 and HNK1 staining (Fig. 3). Render the cells to single cells using accutase followed by replating and maintenance of the cells in Neurobasal media or under alterative culture conditions detailed in Step 4. If desired, cells at day 11 may be further purified for NC lineage using flow cytometric sorting for p75 and HNK1, as described below.
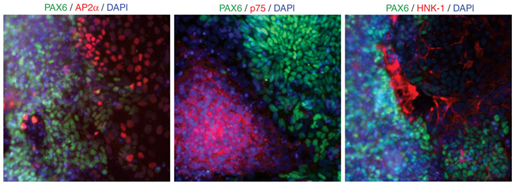
Figure 3: NC cell marker expression using NSB induction. By day 11 in cultures, the NC markers AP2, p75 and HNK1 (all red) can all be found expressed in the non-PAX6 (green)-expressing cells.
2. Isolation of hPSC-derived NC cells using antibody-mediated flow cytometry sorting
- On the day 6 or 7 in P1 culture in the MS5 protocol (see Step 1-1), change medium with accutase (or HBSS) for 20–30 min and harvest the P1 culture with cell scraper.
Critical step Note that we describe this step in detail for cells generated using the MS5 protocol (Step 1-1). However, the same strategy can be adapted for cells generated using the NSB protocol (Step 1-2).
Critical step To determine the presence of NC populations at P1, parallel cultures can be pre-screened by immunocytochemistry. Neuroectodermal marker (Pax6)-positive cells should be interspersed with NC marker (AP2)-positive cells (Figs. 1f and 3). If the goal is to prepare a NC-depleted population, it is possible to manually harvest rosette structures and remove any surrounding cells using a fine needle (27–30 G) attached to 1 ml syringe. - Centrifuge the harvested cells from P1 culture stage for 5 min at 160g at room temperature and aspirate the supernatant. Mechanically triturate cells in PBS containing 2% serum (FBS).
- Label triturated P1 cells (10 million cells per ml) with antibodies (p75, 5–10 μl/ml; HNK1, 10–20 μl/ml) for flow cytometry for 20 min on ice in the dark. Centrifuge for 5 min at 160g at room temperature and aspirate the supernatant.
Critical step The concentration of each lot of antibody may need to be optimized. FACS analysis with appropriate controls is essential to identify optimal antibody dilutions. - Resuspend cells in PBS containing 2% serum (FBS) and label it with appropriate fluorochrome-labelled secondary antibody (1:1000 to 1:500 dilution) for each primary antibody.
- Perform cell sorting by flow cytometry. Collect p75+ and HNK1+ cells (double-positive cells) for further culture (Fig. 4)
Critical step It is essential to have controls of hESC-derived NC cells that are unstained and stained with appropriate secondary antibodies only for defining sorting gates in the FACS machine. As most cells under these conditions are either double positive (p75+ and HNK1+) or double negative, it may be possible to use sorting for single markers (HNK1 or p75) without a major loss of specificity. However, using single markers outside the narrow differentiation window (P1 stage) may result in contamination with other cell types such as CNS neuroblast, cells of placode origin or early mesodermal cells. Dead cells can be excluded for cell sorting by using 7-AAD or DAPI. - Collect double-positive cells in PBS containing 2% serum.
Centrifuge the sorted cells for 5 min at 160g at room temperature and aspirate the supernatant.

Figure 4: Representative image of FACS isolation for NC cells. For flow cytometric isolation, dissociated cells from neural rosettes are sorted for expression of p75 and HNK1.
3-1. Proliferation of sorted NC cells in conventional attaching culture
- Resuspend the pellet by tapping the tube (not pipetting) and plate on culture dishes pre-coated with PO/Lam/FN (Fig. 5, left panel).
Critical step Minimum plating number is 50,000–100,000 cells per cm2 to ensure survival of sorted cells. Viability of sorted cells is high when special precautions are taken to minimize pipetting and other mechanical stress.
Critical step Supplementing the medium with serum or chick embryo extract (as commonly used in primary NC cultures) did not enhance the survival or proliferation of hESC-NC cell cultures under these conditions. For preparing pre-conditioned medium, collect medium from P1 culture (Step 16) and sterilize by 0.22-μm filtration. The pre-conditioned medium can be used by mixing at ratios of 1:1 to 1:6 with fresh NEUROBASAL medium to further enhance survival of sorted cells after plating.
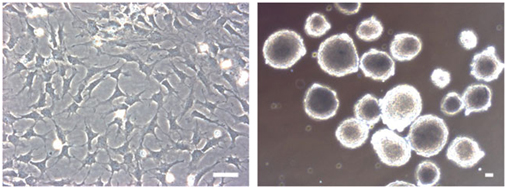
Figure 5: Propagation of sorted NC cells.hESC-derived NC cells are highly enriched in the double-positive (p75+ and HNK1+) population and can be further cultured on PO/Lam/FN-coated plate under serum-free conditions or as sphere on ultra-low attachment plate. Scale bars= 100 μm - Culture the hPSC-NC cells with NEUROBASAL medium supplemented with 10 ng/ml of FGF2 and 10 ng/ml of EGF. Change the media every 2–3 d and passage every 7–8 d. Accutase or HBSS treatment can be used for dissociation of NC cells for splitting.
3-2. Culturing of sorted NC cells in sphere formation
- Resuspend the pellet by tapping the tube (not pipetting) and plate on ultra-low attachment six-well plate in 1,000–10,000 cells per cm2 with 2 ml of NEUROBASAL medium supplemented with 10 ng/ml of FGF2 and 10 ng/ml of EGF. The sphere is typically observed by 24 h after plating (Fig. 5, right panel).
- After every 7 d of culture, dissociate spheres with accutase treatment for 20 min. Filter triturated cells using a 40-μm cell strainer and obtain NC cells. After cell counting using hemacytometer, plate at 1,000–10,000 cells per cm2 into new ultra-low attachment plates in NEUROBASAL medium supplemented with 10 ng/ml of FGF2 and 10 ng/ml of EGF.
4-1. Differentiation of hPSC-derived NC cells toward peripheral neuronal differentiation with hPSC-derived NC cells
- For neuronal differentiation, resuspend dissociated NC cells with Neurobasal medium and plate on culture dishes pre-coated with PO/Lam/FN (20,000 cells per 10 μl drop on pre-dried plate, corresponding to a local density of ~100,000 cells per cm2).
- For the first 2 d, culture hESC-NC cells with NEUROBASAL medium supplemented with 10 ng ml−1 of FGF2 and 10 ng/ml of EGF.
- Afterward, culture cells in Neurobasal medium supplemented with BDNF (10 ng/ml), AA (200 μM), glial cell line-derived neurotrophic factor (10 ng/ml), nerve growth factor (10 ng/ml), Neurotrophin-3 (10 ng/ml) and cyclic AMP (0.5 mM) for 2 weeks. Change the medium including growth factors every 3–4 d (Fig. 6). For improving selective subtype specification toward either sensory or autonomic neuron fate, optimizing growth factor composition, in particular Wnt and BMP signaling factors, is of further consideration.

Figure 6: Specification of hESC-NC cells toward peripheral neurons. hPSC-derived NC cells differentiated toward autonomic neurons (MASH1+/peripherin+) and sensory neurons (Brn3a+/peripherin+). Scale bars=100 µm.
4-2. Schwann cells specification with hPSC-derived NC cells
- After flow cytometric purification at the end of P1 stage (Step 2), culture hESC-NC cells in Neurobasal supplemented with 10 ng/ml of FGF2 and 10 ng/ml of EGF for more than 30 d (Step 3). Depending on the hPSC line used, the optimal time period to maintain cells under FGF2/EGF conditions (Step 3) can vary. The average period is ~60 d. After FGF2/EGF culture, differentiate the ‘aged’ hESC-NC cells along the Schwann cell lineage in Neurobasal medium supplemented with ciliary neurotrophic factor (10 ng/ml), neuregulin (20 ng/ml), FGF2 (10 ng/ml) and cyclic AMP (0.5 mM) for 3 weeks (Fig. 7).
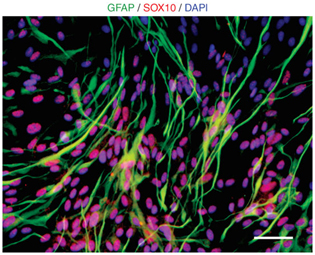
Figure 7: Schwann cell differentiations from hESC-NC cells. Schwann cell fates (GFAP, green; Sox10+, red) are observed from hPSC-derived NC cells. Scale bars=100 µm.Critical step Schwann cell differentiation is not observed during early stages of NC cell culture and we observed glial fibrillary acidic protein-positive (GFAP+) cells only in the aged NC cells cultured for more than 60 d. We propose that the in vitro proliferation period reflect an in vitro maturation (‘aging’) similar to developmental progression in vivo. The developmental window for Schwann cell specification in response to various culture conditions and for individual hPSC lines requires further future studies. It may be important for some of the experiments to formally distinguish GFAP+ Schwann cell precursors generated using this protocol from GFAP+ CNS astrocytes. This can be achieved by double labeling of GFAP+ cells with p75 and/or O4 that are coexpressed in Schwann cells but not in astrocytes. Other Schwann cell makers that can be used for this purpose are Sox10 or myelin basic protein among others.
4-3. Myofibroblast differentiation derived from hPSC-derived NC cells
- Culture the cells for 2–3 weeks in α-MEM medium containing 10% FBS. This should yield hPSC-NC cultures expressing myofibroblast markers (Fig. 8). These cells express a series of cell surface marker such as CD73, Stro-1 and CD146, typically associated with mesenchymal stem cell identity. Indeed, our previous study showed that cells expressing myofibroblast markers in hPSC-NC cultures can readily give rise to adipocytes, chondrocytes, osteocytes and smooth muscle cells, as shown previously for primary cranial NC precursors (Fig. 9).

Figure 8: Representative morphology of myofibroblast cells derived from hESC-NC cells. Scale bars=100 µm.
Figure 9: Adipogenic, chondrogenic and osteogenic differentiation of myofibroblast cells derived from hESC-NC cells. Oil red staining method was used to detect adipocyte. Collagen II staining shows chondrogenic differentiation and alizarin red shows osteogenic cells from hPSC-derived NC cells. Scale bars=50 µm.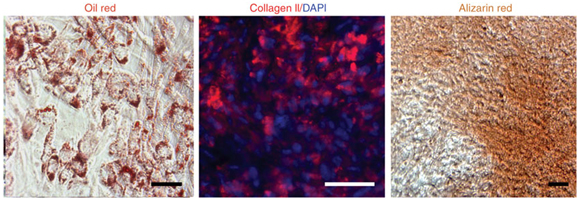
- For adipogenic differentiation, grow hESC-NC-MPCs to confluence, followed by exposure to 1 mM dexamethasone, 10 μg/ml Insulin and 0.5 mM IBMX in α-MEM medium containing 10% FBS for >3 weeks.
- For chondrogenic differentiation, pellet hESC-NC-MPCs for 5 min at 160g at room temperature and take out supernatant without disruption of pellet structure. Then pellet culture can be performed with 10 ng/ml TGFβ-3 and 200 μM AA in α-MEM medium containing 10% FBS for >4 weeks.
- For osteogenic differentiation, plate hESC-NC-MPCs at low density (1 × 103 cells per cm2) on tissue culture-treated dishes in the presence of 10 mM β-glycerol phosphate, 0.1 μM dexamethasone and 200 μM AA in α-MEM medium containing 10% FBS for 3–4 weeks.
Timing
The NC cell differentiation takes ~30 d using the MS5 coculture approach (Step 1-1) and about 12 d using the NSB protocol (Step 1-2). For the isolation and propagation of NC precursor cells (Steps 2–3), an additional period of 1–2 weeks is required. Cultures of cells at the NC precursor stage can be maintained for very extensive time periods (> 4 months) if required. Neuron, Schwann cell and myofibroblast differentiation takes around 3 weeks starting from the appropriate NC precursor cell stage (Step 4). Further differentiation of myofibroblast lineages toward adipogenic, osteogenic and chondrogenic cells will take additional 3–4 weeks (Step 4-3).
Materials:
-
Cell Materials
Mitotically inactivated mouse fetal fibroblasts (PMEF-CF, GlobalStem) hESCs and hiPSCs MS-5 (murine stroma cell line, DSMZ)
-
hES media (1000ml)
Ingredient Amount Company Catalog# DMEM/F12 800ml Life Technology 11330-032 Knockout serum replacement 200ml Life Technology 10829-018 L-Glutamine 5ml Life Technology 21051-016 Pen/Strep 5ml Life Technology 15070-063 MEM-NEAA 10ml Life Technology 11140-050 β –mercaptoethanol 1ml Life Technology 21985-023 FGF-2 4ng/ml R&D system 233-FB-001MG - KSR media (1000ml)
Ingredient Amount Company Catalog# Knockout DMEM 820ml Life Technology 10829-018 Knockout serum replacement 150ml Life Technology 10829-018 L-Glutamine 10ml Life Technology 21051-016 Pen/Strep 10ml Life Technology 15070-063 MEM-NEAA 10ml Life Technology 11140-050 β –mercaptoethanol 1ml Life Technology 21985-023 - Neurobasal media (1000ml)
Ingredient Amount Company Catalog# Neurobasal 950ml Life Technology 21103-049 B27 supplement 20ml Life Technology 12587-010 N2 supplement 10ml Life Technology 17502-048 L-Glutamine 10ml Life Technology 21051-016 Pen/Strep 10ml Life Technology 15070-063 - DMEM with 10% FBS (1000ml)
Ingredient Amount Company Catalog# DMEM 880ml Life Technology 11960-044 FBS 100ml Hyclone 16140-071 L-Glutamine 10ml Life Technology 21051-016 Pen/Strep 10ml Life Technology 15070-063 - α-MEM with 10% FBS (1000ml)
Ingredient Amount Company Catalog# α -MEM 890ml Life Technology 32561-037 FBS 100ml Hyclone 16140-071 Pen/Strep 10ml Life Technology 15070-063 - Reagents
Name Company Catalog# Ca2;/Mg2-free DPBS Life Technology 14190-250 Ca2;/Mg2-free Hanks’ balanced salt solution Life Technology 14170-112 Dispase Life Technology 17105-041 Accutase Innovative Cell Technology AT104 0.05% (wt./vol) trypsin/0.53 mM EDTA Life Technology 25300-054 0.1% Gelatin Millipore ES-006-B Matrigel basement membrane matrix BD 354234 Fibronectin BD 356008 Cultrex Mouse Laminin I R & D Systems 3400-010-01 Poly-L-ornithine hydrobromide Sigma P3655 - Cytokines and Chemicals
- Antibodies
Name Dilution Company Catalog# P75 1:200 Advanced targeting systems AB-N07 Pax6 1:200 Covance PRB-278P AP2 1:200 DSHB 3B5 HNK1 1:300 Sigma C6680 Brn3a 1:200 Millipore AB5945 GFAP 1:300 MP biomedical 8691102 Peripherin 1:200 Santa Cruz sc-7604 MASH1 1:200 BD Pharmingen 556604 - Equipment
Inverted microscope (i.e., Nikon TE or Olympus IX) with fluorescence equipment and digital imaging capture system Horizontal laminar flow hood or a biosafety cabinet with an embedded microscope Biosafety cabinet for cell culture CO2 incubator with controlling and monitoring system for CO2, humidity and temperature Cell culture centrifuge Cell sorting machinery (i.e., MoFlo, Cytomation; FACS Aria, Becton Dickinson; or similar) Glass hemacytometer Cell culture disposables: Tissue culture dishes, multiwell plates, centrifuge tubes, fluorescence-activated cell sorting (FACS) tube, pipettes, pipette tips, etc. Gelatin-coated dishes Matrigel-coated dishes PO/Lam/FN-coated plate (poly-L-ornithine hydrobromide /laminin/ fibronectin)
| Name | Company | Catalog# |
|---|---|---|
| Recombinant mouse Noggin | R&D system | 719-NG |
| Fibroblast growth factor 8 (FGF8) | R&D system | 423-F8 |
| Sonic hedgehog (SHH, C25II substitution) | R&D system | 461-SH |
| Brain-derived neurotrophic factor (BDNF) | R&D system | 248-BD |
| Glial cell line-derived neurotrophic factor (GDNF) | R&D system | 212-GD |
| Wnt3A | R&D system | 1324-WN |
| Bone morphogenetic factor 4 (BMP4) | R&D system | 314-bp |
| Nerve growth factor (NGF) | R&D system | 256-GF |
| Neurotrophin-3 (NT-3) | R&D system | 267-N3 |
| Ciliary neurotrophic factor (CNTF) | R&D system | 257-NT |
| Neuregulin | R&D system | 396-HB |
| Epidermal growth factor (DGF) | R&D system | 236-EG |
| Dickkopf-1 (Dkk-1) | R&D system | 1096-DK |
| TGFβ-3 | R&D system | 243-B3-010 |
| Dibutyryladenosine cyclic monophosphate (dcAMP) | Sigma | |
| Ascorbic acid (AA) | Sigma | A4034 |
| Retinoic acid (RA) | Sigma | R2625 |
| SU5402 | Calbiochem | 572630 |
| Isobutylxanthine (IBMX) | Sigma | I7018-100MG |
| Dexamethasone | Sigma | D2915-100mg |
| β-Glycerol phosphate | Sigma | 50020-100mg |
| SB431542 | Tocris | 1614 |
| Y-27632 dihydrochloride | Tocris | 1254 |
| LDN193189 | Stemgent | 04-0074 |
Troubleshooting:
- No formation of neural rosettes during hESC differentiation (Step 1-2, 11))
- Check the concentration or activity of each cytokines
- Change the batch of MS-5
- Check hESC line for genomic changes or use alternative hESC line
- Change (reduce) the seeding density of hESCs in MS-5 culture
- Check for contamination with Mycoplasma species in hESCs/hiPSCs or MS-5 cells
- Low survival of P1 cells (Step 1-2, 11))
- Check the concentration or activity of each cytokines
- Minimize mechanical stress for rosettes before seeding on PO/Lam-coated dish
- Small percentages of p75+/HNK1+ cells in cell sorting (Step 2)
- Check the dilution of antibodies
- Check settings for cell sorting machine
- Stain P0 or P1 culture before or after cell sorting with antibodies for Pax6, Ap2, p75 and HNK1
- The percentage of NC cells in P1 culture can vary among different hESC lines
- Harvest P1 cells with cell scraper to ensure the detachment of all cells in P1 culture dish
- Low survival of sorted hESC-NC cells (Step 2, 3)
- Use pre-conditioned medium for initial culturing of sorted cells
- Minimize mechanical stress after cell sorting
- Increase plating density after sorting (plate cells at densities ≥ 50,000 to 100,000 cells per cm2)
References:
- Lee, G.et al. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat. Biotechnol. 25, 1468–1475 (2007).
- Lee, G.et al. Modeling pathogenesis and treatment of familial dysautonomia using patient specific iPS cells. Nature 461, 402–406 (2009).
- Chambers, S.M.et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 (2009).
Acknowledgements:
- Thank all the authors of the paper for allowing posting and modification protocols in STEM COREdinates.

