Mesoderm Differentiation: Hematopoiesis
Submitted by: Gordon Keller
Validated by Sunita D’Souza
| Title | Mesoderm Differentiation Hematopoiesis |
| Date Submitted | January 5th 2012 |
| Submitted by - | Kennedy, Marion (mkennedy@uhnresearch.ca) and Keller, Gordon (gkeller@uhnresearch.ca) |
| Adapted from - |
|
| Contributors - | |
| Affiliation(s) - | McEwen Centre for Regenerative Medicine, University Health Network, Toronto, Ontario, Canada.University/ Institute |
*This next section will be on the side and is only relevant for website programmers*
| Score | =Likes/(Likes+Dislikes)+Likes/Downloads or some other algorithm… Suggestions??? I can also come up with something better in due time. |
| Downloads | # |
| "Likes" | # |
| "Dislikes" | # |
| Status | [Validated (>=3, In Progress (=1-2), Not Validated (none)] |
| Validated Core 1 | Name / Institute |
| Validated Core 2 | Name / Institute |
| Validated Core 3 | Name / Institute |
Table of Contents
- Introduction
- FlowChart
- Materials
- Materials and Preparation
- Induction and Differentiation Media
- Images for Hematopoietic Colonies in Methylcellulose Assays
- Protocol
- Acknowledgements
- Contributors
Introduction
The protocol outlined below describes the method for generating hematopoiesis from hESCs, specifically H1 cells. Generally the first hematopoietic progenitors start appearing at T5-7 and are mainly primitive erythroid with a few macrophages. We have defined stages by cell surface markers and colony morphology, in our differentiation cultures, that may reflect the emergence of yolk sac, fetal and definitive hematopoiesis
Flowchart
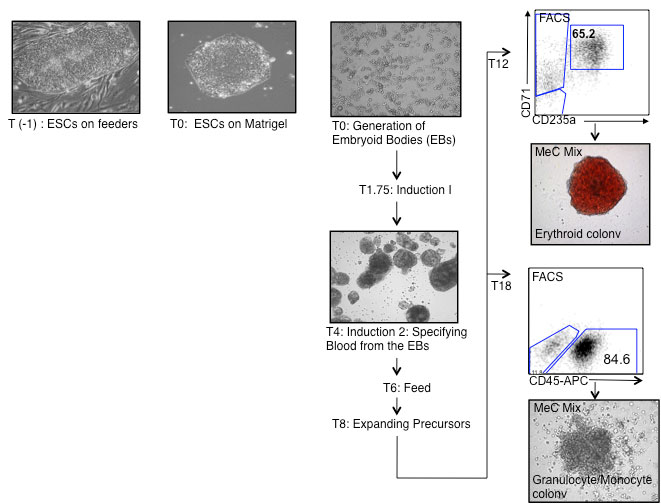
The formation of EBs is the first important step in the differentiation of hESC. This is best achieved by culturing small aggregates of hESCs in miminal amounts of BMP-4 for
24 hours. At this stage, BMP-4 functions to promote the survival of the hESCs.
Materials and Preparation
COLLAGENASE (FOR DISSOCIATION) (SIGMA# C-0130)
Collagenase Type 1 is mainly used for dissociation of the embryoid bodies.
Final Conc. |
For 1 Liter |
||
|---|---|---|---|
Collagenase |
(SIGMA# C-0130) | 0.2% |
2g |
FCS |
|
200mL | |
| PBS (with Ca2+, Mg2 | (Cellgro# 21-030-CM) | 800mL |
- Weigh 1 g of Collagenase Type 1 in the fumehood (inhalation is hazardous) and dissolve in 400 mL of PBS by gently stirring. Check carefully that the collagenase is completely dissolved.
- Add 100 mL FCS
- Filter sterilize (may need more than one filtration apparatus)
- Store at -20ºC as 12 mL aliquots in 15 mL screw cap tubes
- One cycle of freeze-thaw is acceptable.
Note: final concentration is 0.2% or 2 mg/mL in 20% FCS/
L-ASCORBIC ACID (AA) (SIGMA # A-4544)
Prepare a stock solution of 5 mg/mL in cold TC-H2O. Leave on ice and vortex periodically until completely dissolved. Filter sterilize, aliquot and store at -20ºC. Use once and discard
MONOTHIOGLYCEROL (MTG) (SIGMA# M-6145)
The amounts of MTG indicated in our protocols are recommended concentrations. However, it is important to test each new batch of MTG as there is variability between them. MTG should be aliquoted (1 mL) and stored frozen (-20ºC). When aliquots are thawed, they can be used for several experiments and then discarded. Aliquoting of MTG is strongly recommended as it minimizes the amount of oxidation due to repeated opening
TRANSFERRIN (ROCHE# 10 652 202)
The amounts of Transferrin indicated in our protocols are recommended concentrations. However, it is important to test each new batch of transferrin as there is variability between them. It should be aliquoted (2 mL) and stored at 4ºC.
CYTOKINES
All cytokines are stored lyophilized, at -20oC, except Erythropoietin which is stored at 4oC
Cytokine |
Buffer |
Stock Conc. |
|
|---|---|---|---|
hBMP-4 |
(R&D Systems# 314-BP) | H20, 4mM HCL, 0.1%BSA |
10ug/mL |
| hbFGF | (R&D Systems# 234-FSE) | PBS, 0.1%BSA,1mM DTT | 10ug/mL |
| hVEGF | (R&D Systems# 293-VE) | PBS, 0.1%BSA | 5ug/mL |
| hSCF | (R&D Systems# 255-SC) | PBS, 0.1%BSA | 50ug/mL |
| hErythropoietin | Janssen-Ortho Inc (EPREX) | PBS, 0.1%BSA | 2000units/mL |
hIL-6 |
(R&D Systems# 206-IL) | PBS, 0.1%BSA |
5ug/mL |
| hIL-11 | (R&D Systems# 218-IL) | PBS, 0.1%BSA | 5ug/mL |
STEMPRO 34 (Invitrogen# 10639-011)
Stempro 34 is sold as a kit with 2 components. The supplement is kept at -20oC and the liquid media at 4oC. When combined, the media is unstable, therefore, we use it for a maximum of 2 weeks. If not used right away, we store the medium as 50mL aliquots and supplement them as needed. The supplement is frozen as 1.3mL aliquots which is the amount required for 50mL. of medium
Final Conc. |
For 500mL |
||
|---|---|---|---|
STEMPRO 34 Kit |
(Invitrogen 10639-011) |
|
500 mL |
| Penicillin/Streptomycin P/S | (Gibco# 15070-063) | 1% | 5 mL |
- Warm the media, P/S and frozen supplement in a 37oC waterbath for 15 to 20 minutes
- Mix the supplement well with a pipette and add to the warm media along with the P/S.
- Warm the mixture for another 30 minutes and then aliquot into 50mL. (Label the tube that supplement has been added, to avoid confusion.)
- Store for a maximum of 2 weeks
*MATRIGEL (REDUCED FACTOR) (BD# 356 230)
Each batch of matrigel has its own unique levels of endotoxin and protein concentrations. We find that the endotoxin levels should not be higher than 2 endotoxin units/mL and the protein levels should range between 7 to 10 mg/mL. If the protein levels are higher than this you may need to dilute the matrigel more than 1:1. This is determined by observing the hESC colony morphology and the ability of the hESCs to differentiate into the lineage required of them.
Caution: When working with matrigel, all tubes, plates and pipettes should be prechilled, as matrigel solidifies at room temperature.
MATRIGEL1:1 PREPARATION
- Thaw frozen bottles of matrigel on ice overnight in the cold room. We normally thaw 6X5-mL bottles per batch.
- The next day, make a 50% working stock by adding an equal volume of IMDM+P/S to each bottle. Resuspend gently with a pre-chilled 5 mL pipette.
- Leave the bottles on ice all day to allow the matrigel to completely equilibrate with IMDM.
- Pool 3 bottles of 1:1 matrigel (30 mL) into a pre-chilled 50 mL tube. Gently mix with a chilled 10 mL pipette and aliquot.
- Transfer 2.5 mL into pre-chilled and pre-labelled 4-mL snap cap tubes
- Store aliquots at -20ºC
The formation of EBs is the first important step in the differentiation of hESC. This is best achieved by culturing small aggregates of hESCs in the presence of BMP-4 for 24 hours. At this stage, BMP-4 functions to promote the survival of the hESCs.
A. COLLAGENASE B (Roche# 11 088 831 001)
Collagenase B is routinely used for dissociation and passaging of undifferentiated hESCs
Final Conc. |
For 1 Liter |
||
|---|---|---|---|
Collagenase B |
(Sigma# T-4799) | 0.25% |
1 g |
| DMEM/F12 | (Cellgro# 10-092-CV) | 1 mM | 1000 mL |
- Weigh 1 g of Collagenase B in the fume hood as inhalation is hazardous and dissolve in 1 Liter DMEM/F12 +P/S by stirring gently at room temperature for 1 h (or until completely dissolved)
- Filter sterilize and store at -20ºC in 12 mL aliquots
B. DNASE I (VWR, Cat # 80510-412, 10MU)
- Want final concentration to be 1mg/ml
- 10MU X 1X106U ⁄ 1 MU X 1mg ⁄ 65150 U = 153mg
- In the hood transfer powder to a 125 ml bottle
- Bring the volume up to 153 ml with ice cold sterile water
- Let dissolve on ice for 1-2 hours
- Filter and aliquot 1ml/eppendorf
- Store at –20.
- Filter sterilize, aliquot in 1 mL amounts and store frozen at -20ºC
- Use aliquots once and discard excess
C. TRYPSIN-EDTA
Final Conc. |
For 1 Liter |
||
|---|---|---|---|
Trypsin |
(Sigma# T-4799) | 0.25% |
2.5 g |
| EDTA 0.5 M (pH 8) | 1 mM | 2 mL | |
| PBS (without Ca2+, Mg2+) | (Cellgro# 21-031-CM) | 1000 mL |
- Warm to dissolve (15 min, 37ºC), filter sterilize, aliquot and store at -20ºC
- For aggregate formation we use a 1/4 dilution of the above (ie: 10mL of TRYPSIN-EDTA in 30mL of PBS (without Ca2+, Mg2+
D. STOP MEDIUM
Final Conc. |
For 40 mL |
||
|---|---|---|---|
hESC WASH Medium |
50% |
20 mL | |
| WASH Medium 50% 20 mL |
50% | 20 mL | |
| +/- Matrigel (1:1)* | (BD# 356 230) | 1:800 | 100 uL |
E. hESC WASH MEDIUM
Final Conc. |
For 500 mL |
||
|---|---|---|---|
Supplemented DMEM/F12 |
|
475 mL | |
| KnockoutTM Serum Replacement | (Gibco# 10828-028) | 5% | 25 mL |
SUPPLEMENTED DMEM/F12
Final Conc. |
For 500 mL |
||
|---|---|---|---|
DMEM/F12 |
(Cellgro# 10-092-CV) |
|
500 mL |
| Penicillin/Streptomycin | (Gibco# 15070-063) | 1% | 5 mL |
- Make 40 mL aliquots of the hESC WASH MEDIUM
- You can add 100 uL of Matrigel 1:1 to each aliquot when needed
INDUCTION / DIFFERENTIATION MEDIA
F. AGGREGATION MEDIUM
| Stock Conc. | Final Conc. |
per mL |
For 50 ml |
|
|---|---|---|---|---|
STEMPRO 34 |
|
50ml | ||
| Glutamine | 100x | 1% | 10uL | 500ul |
| Transferrin | 300mg/ml | 150ug/mL | 5uL | 250ul |
| Ascorbic Acid | 5mg/ml | 50ng/mL | 10uL | 500ul |
| MTG | 26 λ/2mls | 3uL | 150ul | |
| BMP4* | 10ug/ml | 10ng/mL | 1uL | 50ul |
G. INDUCTION 1 MEDIUM
| Stock Conc. | Final Conc. |
per mL |
For 50 ml |
|
|---|---|---|---|---|
STEMPRO 34 |
|
50ml | ||
| Glutamine | 100x | 1% | 10uL | 500ul |
| Transferrin | 300mg/ml | 150ug/mL | 5uL | 250ul |
| Ascorbic Acid | 5mg/ml | 50ng/mL | 10uL | 500ul |
| MTG | 26 λ/2mls | 3uL | 150ul | |
| BMP4* | 10ug/ml | 10ng/mL | 1uL | 50 |
| ActA** | 10ug/ml | 0.3ng/ml | 0.03uL | 1.5ul |
| bFGF | 10ug/ml | 5ng/ml | 0.5uL | 25ul |
* Bmp4 and ActA**concentration may vary according to lot# or the hES cell line used…these concentrations are for H1 cells.
H. IMDM+10%FCS
Final Conc. |
For 500 mL |
||
|---|---|---|---|
Iscove's Modified Dulbecco's Medium |
(Cellgro# 15-016-CV) |
|
450 mL |
FCS (batch tested) |
(Gibco# 10828-028 ) | 10% |
50 mL |
I. INDUCTION 2 MEDIUM
| Stock Conc. | Final Conc. |
per mL |
For 50 ml |
|
|---|---|---|---|---|
STEMPRO 34 |
|
50ml | ||
| Glutamine | 100x | 1% | 10uL | 500ul |
| Transferrin | 300mg/ml | 150ug/mL | 5uL | 250ul |
| Ascorbic Acid | 5mg/ml | 50ng/mL | 10uL | 500ul |
| MTG | 26 λ/2mls | 3uL | 150ul | |
| VEGF | 5ug/ml | 10ng/mL | 1uL | 100ul |
| Dkk | 50ug/ml | 150ng/mL | 3uL | 150ul |
| bFGF | 10ug/ml | 5ng/ml | 0.5uL | 25ul |
J. INDUCTION 2 MEDIUM (Feed T6)
| Stock Conc. | Final Conc. |
per mL |
For 50 ml |
|
|---|---|---|---|---|
STEMPRO 34 |
|
50ml | ||
| Glutamine | 100x | 1% | 10uL | 250ul |
| Transferrin | 300mg/ml | 150ug/mL | 5uL | 100ul |
| Ascorbic Acid | 5mg/ml | 50ng/mL | 10uL | 500ul |
| MTG | 26 λ/2mls | 3uL | 150ul | |
| VEGF | 5ug/ml | 10ng/mL | 2uL | 100ul |
| bFGF | 10ug/ml | 5ng/mL | 0.5uL | 50ul |
| hIL6 | 10ug/ml | 10ng/ml | 2ul | 100ul |
| hSCF | 50ug/ml | 50ng/ml | 2ul | 100ul |
| hIL-11 | 5ug/ml | 5ng/ml | 2ul | 100ul |
| hEPO | 2000units/ml | 2units/ml | 2ul | 100ul |
| hIGF-1 | 25ug/ml | 25ng/ml | 2ul | 100ul |
Bold type indicates a 2x concentration of the cytokine as the media will be diluted by 1/2
K. HEMATOPOIETIC EXPANSION MEDIUM
| Stock Conc. | Final Conc. |
per mL |
For 50 ml |
|
|---|---|---|---|---|
STEMPRO 34 |
|
50ml | ||
| Glutamine | 100x | 1% | 10uL | 500ul |
| Transferrin | 300mg/ml | 150ug/mL | 5uL | 250ul |
| Ascorbic Acid | 5mg/ml | 25ng/mL | 5uL | 250ul |
| MTG | 26 λ/2mls | 2uL | 100ul | |
| hTPO | 10ug/ml | 50ng/mL | 5uL | 250ul |
| hIL-3 | 10ug/ml | 50ng/mL | 5uL | 250ul |
| hFLT3L | 10ug/ml | 20ng/ml | 2ul | 100ul |
| hSCF | 50ug/ml | 100ng/ml | 2ul | 100ul |
| hIL-11 | 5ug/ml | 5ng/ml | 1ul | 50ul |
| hEPO | 2000units/ml | 2units/ml | 1ul | 50ul |
| hIGF-1 | 25ug/ml | 25ng/ml | 1ul | 50ul |
HEMATOPOIETIC MEC MIX
Mec Mix |
||||||||
|---|---|---|---|---|---|---|---|---|
Reagent |
Stock/ml | Final |
10ml | 14ml | 18ml | 24ml | 32ml | 45ml |
| MEC | 100% | 55% | 5.5ml | 7.7ml | 10ml | 13ml | 17.6ml | 25ml |
| PDS | 100% | 15% | 1.5ml | 2.1ml | 2.7ml | 3.6ml | 4.8ml | 6.8ml |
| PFHM-11 | 100%l | 5% | 0.5ml | 0.7ml | 0.9ml | 1.2ml | 1.6ml | 1.3ml |
| Glutamine | 100% | 1x | 100ul | 140ul | 180ul | 240ul | 320ul | 450ul |
| Transferrin | 30mg | 150ug/ml | 50ul | 70ul | 90ul | 120ul | 160ul | 275ul |
| TPO | 10ug | 50ng/mL | 50ul | 70ul | 90ul | 120ul | 160ul | 225ul |
| IL-3 | 10ug | 50ng/ml | 50ul | 70ul | 90ul | 120ul | 160ul | 225ul |
| VEGF | 5ug | 10ng/ml | 20ul | 28ul | 36ul | 48ul | 64ul | 90ul |
| SCF | 50ug | 100ng/ml | 20ul | 28ul | 36ul | 48ul | 64ul | 90ul |
| EPO | 2000units/ml | 4units | 20ul | 28ul | 36ul | 48ul | 64ul | 90ul |
| IL-6 | 5ug/ml | 10ng | 20ul | 28ul | 36ul | 48ul | 64ul | 90ul |
| IGF-1 | 25ug | 50ng | 20ul | 28ul | 36ul | 48ul | 64ul | 90ul |
| IL-11 | 5ug | 5ng | 10ul | 14ul | 18ul | 24ul | 32ul | 45ul |
| GM-CSF | 1ug | 1ng | 10ul | 14ul | 18ul | 24ul | 32ul | 45ul |
| IMDM | 100% | 2.2ml | 3ml | 3.9ml | 5.2ml | 7ml | 9.7ml |
Protocol
T0: Generation of embryoid bodies (EBs)
- Remove the medium from hES cells that have been feeder depleted on matrigel coated plates for 24-48 hours(see hES cell maintainance protocol)
- To each well, add 1mL of COLLAGENASE B containing 10uL/mL DNase (A,B), for 20 min. and then aspirate. The cells should still be attached.
- Add 1mL of 1/4 TRYPSIN-EDTA (C) to the wells and then watch carefully for 1-3min. The cells will separate from each other, but should not lift from the plate. (Note; each cell line has a different time requirement so one should continuously monitor the wells.
- Remove the trypsin from the well and stop the reaction with 1mL of STOP MEDIUM +MATRIGEL (D) containing 10uL/mL DNase . Scrape gently with a cell scraper. The ES cells should lift as clusters into the medium.
- Add 1ml of hESC WASH MEDIUM +MATRIGEL (E) to each well and resuspend with a 2mL pipette 3-5x, until the clusters are 10-20 cells in size. Transfer to a 15ml tube containing 4ml of hESC WASH MEDIUM +MATRIGEL (usually 3 wells per tube)
- Also ,at this time, we completely trypsinize one well of the matrigel hESCs to single cells, to get an accurate cell count for the experiment. The average cell count per starting matrigel well, using this protocal, is 5x10^5 to 1x10^6.
- Spin at 800 rpm, aspirate and resuspend each tube with 2mL of AGGREGATION MEDIUM (F) using a 2mL pipette (resuspend gently 2-3 times). To calculate the amount of AGGREGATION MEDIUM required for the experiment you will need 2mls for every 5x10^5 to 1x10^6 cells harvested. Adjust the final volume of each tube appropriately.
- With a 5mL pipette, evenly distribute 2 mL of aggregates into each well of a 6 well Locluster plate (Costar#3071). Incubate for 24 hours at 37°C in an 5%CO2,5%O2 Incubator.
T1: Feeding Aggregates and expansion
Add 2mls of AGGREGATION MEDIUM (F) supplemented with 10ng/ml bFGF to the cultures. The final concentration of bFGF in the well will be 5ng/ml.
-
Please note low concentration of BMP4 (1-2ng/ml) at D0, followed by 10ng of BMP4 at D1-D4 and 4-5ng of ActA from D1-D4 led to a drastic increase in the amount of erythroid progenitors. All other cytokines are as described.
- Whereas 10ng/ml of BMP4 at D0, followed by 10ng of BMP4 from D1-D4 and 0.5 ng of ActA from D1-D4 led to a drastic increase in the amount of myeloid progenitors. All other cytokines are as described.
T1.75: Induction 1
-
There will be some debris in the cultures. We separate the aggregates from the debris by harvesting 3 wells into one 15ml round bottomed tube and then allowing them to settle for 30min in an 37°C,5%CO2,5%O2 incubator
-
While the aggregates are settling, make INDUCTION 1 MEDIUM (G) so that there will be 2mL for each well harvested. After the aggregates have settled, aspirate the supernatant and then add INDUCTION 1 MEDIUM . Dispense the aggregates into a new 6 well Locluster plate at 2mL per well. Distribute the aggregates evenly.
- Incubate as above until T3-4
Notes
Mesoderm induction and hemangioblast specification in the EBs should be evaluated by flow cytometric analysis, monitoring the cells for expression of KDR and CD117 (c-kit). As each hESC line has its own unique kinetics, it is best to define the hemangioblast stage based on the profile seen below, rather than by time in culture.
The hemangioblast stage is defined by the appearance of a population that expresses medium levels of KDR and are CD117neg . EBs at this stage also contain a KDRneg CD117pos population. This profile is detected in H1-derived EBs at day 3 and in HES2-derived EBs at day 4. The window for hemangioblast development is narrow and occurs before CD31 is expressed on the cells.
T3-4: Harvest for staining
- Settle1 well of EBs in a round bottomed tube for 15minutes. Aspirate the medium and add 1ml of TRYPSIN-EDTA. Incubate at 37°C in a waterbath for 5 minutes and then stop the reaction with 0.5ml of STOP MEDIUM+dnase.
- Make to single cells by passing the EBs 4-6x through a syringe bearing a 20 Guage needle and wash with IMDM+10%FCS (H).
- Spin for 5min at 1000 RPM, aspirate and resuspend in IMDM+FCS (usually 500ul per well harvested). Pass the cells through a 70micron filter to remove any clumps that are still remaining. You should recover 5x10^5-1x10^6 cells per well of EBs harvested
Facs Profiles

T4-8: Induction 2: Specifying Blood from the EBs
- Pipette the EBs into a round bottomed 15ml tube (1-3 wells per tube). Spin for 5 min at 500RPM. Carefully aspirate the media and then add 4ml of Stempro media. Let the EBs settle for 20 min and aspirate again. This step allows for 2 washes to remove the previous cytokines, as well as, any debris floating in the supernatant.
- Resuspend the EBs in INDUCTION 2 MEDIA (I) at 2mls per well harvested. This media contains VEGF and FGF at low concentrations which is necessary for expanding the earliest blood precursors. Incubate at 37°C in an 5%CO2,5%O2 Incubator.
- Feed the cultures at T6 with 2ml of the INDUCTION 2 MEDIA (J) with supplemental cytokines which are in BOLD and added at a 2x concentration.
Notes
If one monitors the cultures at this time with FACS, the KDR/CD117 and KDR/CD31 profiles should be the same as the Post-Hemangioblast profiles above.
T8: Expanding Precursors
Settle and wash the EBs as above at T4
Resuspend and distribute as above in HEMATOPOIETIC EXPANSION MEDIUM (K) At this time cocktails of cytokines may vary as well as culture methods according to the lineage one desires. For general expansion of hematopoietic lineages we maintain our cultures in suspension changing the media every 4 days. The cultures at this time can be grown in ambient oxygen conditions at 37° and 5%CO2
Harvest to single cells for FACs, Sort, Counts or Reaggregations
Dissociation T1-8 (Trypsin-EDTA)
- Harvest each group of 2 dishes into a 2059 tube and let the EBs settle
- Remove supernat.and add 2mls of Cellgro Trypsin
- Incubate for 5-8min in 37degree water bath
- Stop trypsin with FCS1:1+dnase 30ul/ml
- Syringe 6x with a 20Guage needle and then add 7mls of wash media
- Spin for 5min at 1100rpms
- Resuspend in 1ml of wash media and filter through blue cap of
2035 tube
Dissociation T8+(Collagenase)
- Harvest each group of 2 dishes into a 2059 tube and let the EBs settle
- If there are hematopoietic clusters in your supernatant collect the sup., keep on ice during the dissociation step and add back to your cells at step 9
- Remove supernat.and add 2mls of Cellgro Trypsin
- Incubate for 5-8min in 37degree water bath
- Stop trypsin with FCS1:1+dnase 30ul/ml
- Syringe 6x with a 20Guage needle and then add 7mls of wash media
- Spin at 1200rpm and resuspend in 1ml/well Collogenase Type 1 (L)
- Incubate at 37 degrees for 60min
- Syringe 6x with a 20Guage needle, add your supernatant cells and wash media.
- Spin at 1200rpm and resuspend in 1ml.
- Count and use in your assays
IMAGES OF HEMATOPOIETIC COLONIES IN METHYLCELLUOSE COLONIES
MAST COLONY – 20X

MAST COLONY – 10X

MAC/MONO – 10X

MAC/MONO – 10X

MACROPHAGE – 10X

MACROPHAGE – 20X
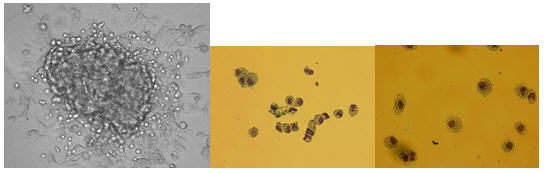
ERY-MYELO – 10X

MACROPHAGE – 10X

ERYTHROID – 10X
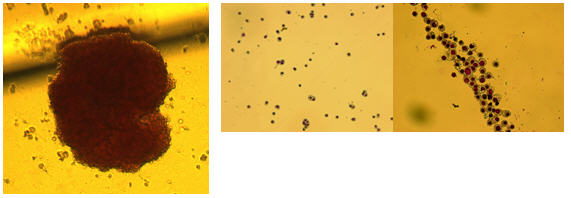
Granulocyte/Macrophage

Acknowledgements
References
Blood. 2010 Apr 8;115(14):2769-76. Epub 2010 Jan 11.
Directed differentiation of hematopoietic precursors and functional osteoclasts from human ES and iPS cells.
Grigoriadis AE, Kennedy M, Bozec A, Brunton F, Stenbeck G, Park IH , Wagner EF, Keller GM.
Department of Craniofacial Development and Orthodontics, Guy's Hospital, King's College London, London, UK. agi.grigoriadis@kcl.ac.uk
- Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures.
Kennedy M, D'Souza SL, Lynch-Kattman M, Schwantz S, Keller G. Blood. 2007 Apr 1;109(7):2679-87.

