 |
||
| http://ghr.nlm.nih.gov/ A service of the U.S. National Library of Medicine® | ||
The official name of this gene is “K(lysine) acetyltransferase 6B.”
KAT6B is the gene's official symbol. The KAT6B gene is also known by other names, listed below.
The KAT6B gene provides instructions for making a type of enzyme called a histone acetyltransferase. These enzymes modify histones, which are structural proteins that attach (bind) to DNA and give chromosomes their shape. By adding a small molecule called an acetyl group to particular locations on histones, histone acetyltransferases control the activity of certain genes.
Little is known about the function of the histone acetyltransferase produced from the KAT6B gene. It is active in cells and tissues throughout the body, where it interacts with many other proteins. It appears to regulate genes that are important for early development, including development of the skeleton and nervous system.
The KAT6B gene belongs to a family of genes called chromatin-modifying enzymes (chromatin-modifying enzymes).
A gene family is a group of genes that share important characteristics. Classifying individual genes into families helps researchers describe how genes are related to each other. For more information, see What are gene families? (http://ghr.nlm.nih.gov/handbook/howgeneswork/genefamilies) in the Handbook.
At least eight mutations in the KAT6B gene have been identified in people with genitopatellar syndrome, a rare condition characterized by genital abnormalities, missing or underdeveloped kneecaps (patellae), intellectual disability, and abnormalities affecting other parts of the body. The mutations that cause genitopatellar syndrome occur near the end of the KAT6B gene in a region known as exon 18. These mutations lead to the production of a shortened histone acetyltransferase enzyme. Researchers suspect that the shortened enzyme may function differently than the full-length version, altering the regulation of various genes during early development. Because the altered enzyme takes on a different function, these mutations are described as "gain-of-function." However, it is unclear how these changes lead to the specific features of genitopatellar syndrome.
More than 10 mutations in the KAT6B gene have been found to cause the Say-Barber-Biesecker-Young-Simpson (SBBYS) variant of Ohdo syndrome. This condition has signs and symptoms that overlap with those of genitopatellar syndrome (described above), although some of the specific developmental abnormalities differ between the two conditions. Mutations that cause the SBBYS variant of Ohdo syndrome have been identified throughout the KAT6B gene, although many of them occur in exon 18. Studies suggest that these mutations likely prevent the production of functional histone acetyltransferase from one copy of the KAT6B gene in each cell. A shortage of this enzyme impairs the regulation of various genes during early development. Because these mutations lead to a reduction in the enzyme, they are described as "loss-of-function." However, it is unclear how these changes lead to the specific features of the condition.
Genetic changes involving the KAT6B gene have been associated with certain types of cancer. These mutations are somatic, which means they are acquired during a person's lifetime and are present only in certain cells. The genetic changes are chromosomal rearrangements (translocations) that disrupt the region of chromosome 10 containing the KAT6B gene. Researchers have found a translocation that attaches this region of chromosome 10 to part of chromosome 16 in some people with a cancer of blood-forming cells called acute myeloid leukemia (AML). This translocation has also been identified in some people with therapy-related myelodysplastic syndrome, a blood disorder that can occur after a person has undergone chemotherapy for another form of cancer.
It is unclear how translocations involving the KAT6B gene are related to the development of cancer. These changes likely alter histone modification, which could prevent normal regulation of gene activity. Impaired gene regulation may contribute to the growth of cancers by allowing abnormal cells to grow and divide uncontrollably.
Somatic changes involving the KAT6B gene have also been identified in some people with uterine leiomyomas, which are noncancerous growths in the uterus that are also known as uterine fibroids. Uterine leiomyomas are common in adult women. These growths can cause pelvic pain and abnormal bleeding, and, in some cases, lead to an inability to have biological children (infertility). The genetic change associated with uterine leiomyomas is a translocation between the region of chromosome 10 containing the KAT6B gene and a particular region of chromosome 17. It is unclear how this translocation is related to tumor development. Changes in histone modification that impair normal gene regulation may allow certain cells to divide in an uncontrolled way, leading to the growth of a tumor.
Cytogenetic Location: 10q22.2
Molecular Location on chromosome 10: base pairs 76,586,170 to 76,792,379
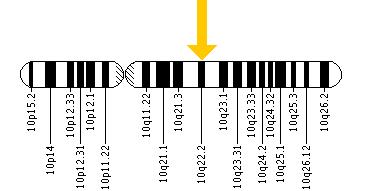
The KAT6B gene is located on the long (q) arm of chromosome 10 at position 22.2.
More precisely, the KAT6B gene is located from base pair 76,586,170 to base pair 76,792,379 on chromosome 10.
See How do geneticists indicate the location of a gene? (http://ghr.nlm.nih.gov/handbook/howgeneswork/genelocation) in the Handbook.
You and your healthcare professional may find the following resources about KAT6B helpful.
You may also be interested in these resources, which are designed for genetics professionals and researchers.
See How are genetic conditions and genes named? (http://ghr.nlm.nih.gov/handbook/mutationsanddisorders/naming) in the Handbook.
acute ; acute myeloid leukemia ; AML ; cancer ; cell ; chemotherapy ; chromosome ; DNA ; domain ; enzyme ; exon ; gene ; gene regulation ; histone ; infertility ; leukemia ; molecule ; mutation ; myelodysplastic syndrome ; myeloid ; nervous system ; patella ; protein ; rearrangement ; sign ; symptom ; syndrome ; tissue ; translocation ; tumor
You may find definitions for these and many other terms in the Genetics Home Reference Glossary (http://ghr.nlm.nih.gov/glossary).
The resources on this site should not be used as a substitute for professional medical care or advice. Users seeking information about a personal genetic disease, syndrome, or condition should consult with a qualified healthcare professional. See How can I find a genetics professional in my area? (http://ghr.nlm.nih.gov/handbook/consult/findingprofessional) in the Handbook.