What is structural biology?
Structural biology is a field of science focused on understanding the structures of biological molecules in order to learn more about how they function and interact. These molecules include DNA, RNA, proteins, carbohydrates (sugars) and lipids (fats). Structural biologists are particularly interested in proteins because they do so much of the work in the body.
What are proteins?
Proteins are molecules that play impnortant roles in virtually every activity in the body. They form hair and fingernails, carry oxygen in the blood, enable muscle movement and much more.
What are proteins made of?
Proteins are made of long strands of small molecules called amino acids. There are 20 different amino acids in nature, and each protein contains a unique combination of a few dozen to many thousands of them. Some proteins are made up of multiple amino acid strands that wind together in the completed protein.
How does a protein get its shape?

Even though a protein is made up of a string of amino acids, it does not remain as a straight chain. The strand twists, bends and folds into a specific shape. Some sections are recognizable “motifs”: corkscrew-like coils called alpha helices and flat sections called beta sheets.
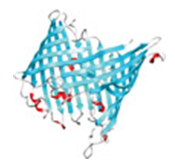

Although researchers can easily determine a protein’s amino acid sequence, they haven’t completely cracked the code that governs how proteins fold. This so-called “protein folding problem” remains one of the greatest challenges in structural biology. If scientists could solve it, they could better understand how proteins misfold and malfunction in diseases like Alzheimer’s and cystic fibrosis. This might enable them to design new treatments.
Why does a protein’s shape matter?
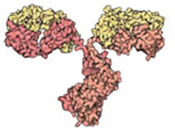
A protein’s structure enables it to perform its task. For instance, the Y shape of antibodies helps these immune system proteins bind to foreign molecules like bacteria or viruses while also drawing in other immune system molecules. The donut shape of DNA polymerase III helps this protein form a ring around DNA to copy its genetic information. The grooves or pockets of proteins called enzymes help them hold onto other molecules to speed chemical reactions.
What kinds of proteins are there?
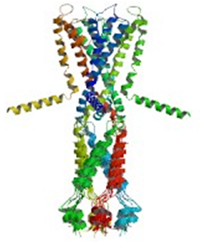
Many proteins are enzymes that facilitate biochemical reactions. Others have structural or mechanical functions that provide scaffolding to maintain cell shape or help cells move. Another type forms channels that help substances like sodium or potassium pass into or out of the cell through its outer membrane. Proteins that are embedded in the cell’s membranes are among the most challenging to study, partly because they tend to fall apart or stick together when scientists try to work with them.
What have we learned by studying the structures of molecules?
Because structural biology enables researchers to see what molecules look like in three dimensions, it has helped scientists understand how the thousands of different molecules in each of our cells work together to keep us healthy. Structural studies have also provided insights on how misshapen molecules make us sick and have suggested new treatment strategies for many diseases.
How do scientists use structure to develop new drugs?
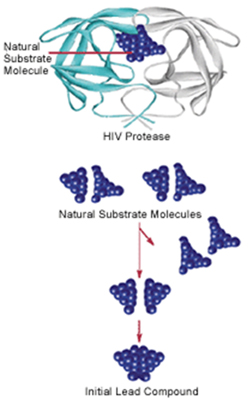
Drugs work by either blocking or stimulating the activity of specific proteins in the body, but knowing how to affect a protein’s activity can be tricky. Using an approach called structure-based drug design, scientists can use the structure of a protein as a template for identifying new medicines. They start with a computerized structural model of the protein bound to a natural molecule, called a substrate, which “unlocks” a biological action. Here, the protein is the lock, and the substrate is the key. If scientists want to turn off an enzyme, for example, they would try to design a molecule that would block the lock. They also can search for a molecule that fits the lock more tightly than an existing drug molecule to create a more effective medicine.
What’s an example of a medicine developed using structure-based drug design?
Structure-based drug design was used in the development of some anti-HIV drugs. HIV protease is an enzyme essential in the virus’s life cycle, and knowing its structure allowed researchers to determine the kinds of molecules (“keys”) that could block the active site (“lock”) and stop HIV protease from functioning. Scientists were able to use computer models to determine which molecules would fit and halt virus production. This work led to new medicines called protease inhibitors.
How do scientists determine protein structures?
The two most common methods for determining protein structures are X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy. A third method, cryo-electron microscopy, is also used to determine the shapes of large structures like molecular complexes (groups of proteins that combine and function as a unit), viruses or organelles.
What is X-ray crystallography and how does it work?

X-ray crystallography shoots a beam of X-rays through a tiny solid crystal made up of trillions of identical protein molecules. The crystal scatters the X-rays onto an electronic detector, similar to those that capture images in a digital camera. The intensities of the scattered X-rays are fed into a computer, which calculates the position of each atom in the crystallized molecule. The result is a three-dimensional, digital image of the molecule. More than 85 percent of known protein structures have been determined using this method.
What is NMR spectroscopy and how does it work?

NMR spectroscopy relies on the interaction between an applied magnetic field and the natural magnets—the nuclei of certain atoms—inside proteins. When scientists place a protein in an NMR machine, the magnets in the protein’s atoms line up with a big magnet inside the machine. The researchers then blast the sample with a series of split-second radio-wave pulses and observe how the protein’s magnets respond. By doing several sets of experiments and combining the data, scientists can put together a complete picture of the protein. NMR spectroscopy is limited to smaller proteins than X-ray crystallography. An advantage of NMR spectroscopy over X-ray crystallography is that it uses proteins in a watery solution, which avoids the often tricky step of making crystals from proteins.
What is cryo-electron microscopy and how does it work?
In cryo-electron microscopy, a cell, virus, molecular complex or other structure is frozen so rapidly that water molecules do not have time to form crystals. This preserves the sample in its natural state. Scientists expose the frozen sample to a beam of electrons in an electron microscope, creating a two-dimensional projection of the sample on a digital detector placed beneath it. By computationally averaging many orientations of the sample, scientists generate a three-dimensional model of its structure. While the model is not as detailed as those obtained using X-ray crystallography or NMR spectroscopy, the overall shape can provide valuable insights. For example, visualizing the arrangement of molecules on viral surfaces could help scientists understand how vaccines work and how to design more effective ones.
What does the future hold for structural biology?
Technical advances made over several decades have enabled researchers to determine the structures of increasingly larger molecules and complexes, including the protein-making ribosome—a molecular behemoth composed of several RNA molecules and dozens of proteins and the target of many antibiotics. Today, scientists are using high-throughput methods to determine protein structures more quickly than ever before. They are also using computational techniques to predict three-dimensional structures of proteins of unknown structure and to design new proteins with useful functions. This work will continue to increase our understanding of the diverse roles molecules play in biology and to spur advances in medicine.
Learn more:
The Structures of Life
Computing Structural Biology from Computing Life
For Proteins, Form Shapes Function
NIGMS is a part of the National Institutes of Health that supports basic research to increase our understanding of life processes and lay the foundation for advances in disease diagnosis, treatment and prevention. For more information on the Institute's research and training programs, see http://www.nigms.nih.gov.
Content reviewed November 2012