The human kinome (part 1)
Update Nov 27, 2012: The entire source code for this tool is available here.
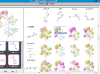
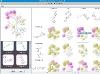
Kinases are an important class of enzymes that are therapeutically relevant for a wide range of indications such as cancer and neurodegenerative diseases. Given that there are roughly 500 different kinase proteins, selectivity—and in some cases polypharmacology—is perhaps one of the most important parameters used during lead optimization. Here the lead compound is subjected to a panel of kinase assays and the results are often projected onto the kinome dendrogram. Such visualizations provide an effective means to evaluate the selectivity profile of lead compounds. Recently, we develop a self-contained kinome prototype tool for this very purpose. At the moment the tool assumes that kinase activities are expressed in μM. Here is an example of the input format for the tool. As always, we welcome suggestions and/or feedback.
Although being able to robustly project data onto the kinome dendrogram is an important step in evaluating selectivity, a more useful scenario is to consider the underlying kinase profile in a broader context of known/published results. In part 2 of this post, we discuss the integration of our kinome viewer with the kinase subset of the ChEMBL database. Please stay tuned.