Understanding Critical Protein Structures May Speed Drug Development
Identifying new compounds to treat heart disease, cancer, or other inflammatory
and immune-related disorders may get easier using a precisely tailored application
of solid state nuclear magnetic resonance (NMR) spectroscopy–which can reveal
a protein’s entire 3-D structure within its natural surroundings. Researchers
funded by the National Institute of Biomedical Imaging and Bioengineering (NIBIB),
part of the National Institutes of Health (NIH), who developed the solid state NMR
techniques describe their use of this technology to determine the structure of a
receptor commonly targeted by breast cancer medications in the October 21 online
issue of Nature.
“The major advantage of solid state NMR over other methods of structural analysis
is knowing that the structure reflects what’s actually there, rather than
artifacts created while extracting the protein for study,” said Alan McLaughlin,
Ph.D., Director of the NIBIB Division of Applied Science and Technology. “Having
such reliable and high resolution structures may lead to more precise compounds
that bind tightly to their intended receptors. This is a very significant advance.”
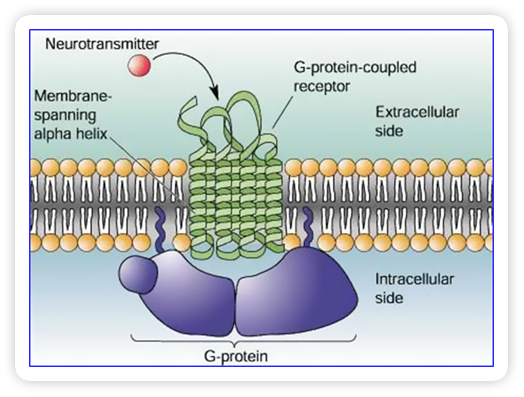
Over half of all currently available medications target G-protein-coupled receptors
(GPCRs), a large family of proteins that span the membrane of a cell, with receptors
outside the cell and G protein binding sites inside. This feature allows specific
substances outside the cell to affect functions inside the cell, making GPCRs key
treatment targets. However, this same membrane-spanning feature makes their structure
hard to study.
Understanding the structure of a particular binding site allows researchers to work
backwards and design a compound that can interact with that site. Standard approaches
such as x-ray crystallography can determine the structure of proteins, but only
after they have been crystallized. In some cases these crystallographic approaches
can greatly shorten the treatment development timelines, for example with HIV. But
in the case of GPCRs the process of creating crystals, which involves adding chemical
detergents or extra proteins, could change the GPCR structure.
To address these issues, Stan Opella, Ph.D., Professor of Chemistry and Biochemistry
at the University of California at San Diego, and colleagues combined and adapted
a number of NMR techniques, collectively referred to as solid state NMR spectroscopy,
to obtain high resolution structural data of an intact, large membrane protein in
its native environment. Solid state NMR approaches thus complement recent x-ray
crystallography studies of GCPRs.
Specifically, the researchers were able to determine the structure of a GPCR called
CXCR1, which is one of two major receptors for the CXC cytokine called interleukin-8
(IL 8). Cytokines are secreted by immune cells that carry important functional signals
to other immune cells.
When attached to the CXCR1 receptor, IL-8 influences immune and inflammatory responses.
CXCR1 and IL-8 have been associated with tumor growth, heart disease, and many other
inflammatory and immune-related disorders.
Dr. Opella’s solid state NMR approach could be used to determine the structure
of other GPCRs as well as many other proteins in the membranes of cells.
“The support we got from NIBIB allowed us to do the basic fundamental NMR
research and focus our efforts on the purification and refolding of a single GPCR
to get an active receptor. It was essential for the ultimate success of this project,”
said Opella.
The significance of this area of research was recently acknowledged when the Nobel
Prize in Chemistry was awarded to Robert Lefkowitz and Brian Kobilka for their initial
work with GPCRs, principally using x-ray crystallography. “Dr. Opella’s
work extends the early observations of GPCR structure to the next level,”
said Dr. Roderic Pettigrew, Director of NIBIB. “It will be exciting to see
where the development of this process that allows the study on unaltered proteins
takes us.”
In addition to NIBIB, this study was supported by the National Institute of General
Medical Sciences and National Institute for Allergy and Infectious Diseases, both
part of NIH, as well as the Cambridge Isotope Laboratories, Swiss National Science
Foundation, and Novartis Foundation.
-- Jessica Meade and Karin Lee
 More Highlight
on Health articles
More Highlight
on Health articles
Last Updated On 11/13/2012