News: 2008
October
Transformative R01 Round Table Discussion: 3-D Tissue Models
The NIH has issued a Roadmap initiative calling for “Transformative R01”
applications (http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-08-029.html).
Since this will be a new program, we anticipate that the research community will
have many questions. NIH will host a meeting to help establish a dialog and begin
to address some of these questions for one of the “areas of highlighted need
that have been identified through an NIH strategic planning process” in this
initiative, specifically the generation of complex, 3-dimensional tissue models.
The meeting will be a “Round Table Discussion” hosted by Dr. Alan Krensky
(Director, NIH Office of Portfolio Analysis and Strategic Initiatives) and will
include about a dozen discussants with experience in the area of in vitro
engineered tissues. The discussion will be held on the NIH campus, Building 31/Conference
Room 10 on Friday, October 24, 2008, 8:30 AM – 12:30 PM. The broad objective
will be to illuminate potential transformative research for the field—to distinguish
between incremental progress and work that will truly disrupt current paradigms,
or create new ones where none exist. This will be an open meeting and will also
be broadcast live from the NIH Videocast web site (http://videocast.nih.gov/).
There is also a link to the meeting from the NIH Roadmap web site at: http://nihroadmap.nih.gov.
Invited Discussants:
- Linda Griffith (Massachusetts Institute of Technology)
- Jonathan Garlick (Tufts University)
- Anna-Katerina Hadjantonakis (Sloan-Kettering Institute)
- Christy Haynes (University of Minnesota)
- Meenhard Herlyn (Wistar Institute)
- Karen Hirschi (Baylor College of Medicine)
- Michelle LaPlaca (Georgia Institute of Technology)
- Milan Mrksich (University of Chicago)
- David Mooney (Harvard University)
- Cheryl Nickerson (Arizona State University)
- Shuichi Takayama (University of Michigan)
- Herman Vandenburgh (Brown University)
- Gordana Vunjak-Novakovic (Columbia University)
Back to Top
July
A Sensible Censor for Sharing Medical Records
MIT-developed Software Helps Protect Patient Privacy
Elizabeth A. Thomson, MIT News Office
July 23, 2008
Newly developed MIT software will help to allay patients' fears about who has
access to their confidential records, facilitating the use of that data for medical
research.
In the July 24 issue of the journal BMC Medical Informatics and Decision Making,
a team of MIT researchers describes a computer program capable of automatically
deleting details from medical records that may identify patients, while leaving
important medical information intact.
Patient records that are to be shared within the research community must have any
identifying information removed, according to the U.S. Health Insurance Portability
and Accountability Act (HIPAA). However, manual removal of identifying information
is prohibitively expensive, time consuming and prone to error-constraints that have
prompted considerable research toward developing automated techniques for "de-identifying"
medical records.
The MIT team aimed to solve this problem. "We've developed a free and open-source
software package to allow researchers to accurately de-identify text in medical
records in a HIPAA-compliant manner," said Gari D. Clifford, a principal research
scientist in the Harvard-MIT Division of Health Sciences and Technology (HST) who
led the work with Principal Investigator Roger G. Mark, a professor in HST and MIT's
Department of Electrical Engineering and Computer Science.
According to Dr. Zohara Cohen, program director at the National Institute of Biomedical
Imaging and Bioengineering, sponsor of the work, the information in patients'
medical records is a "largely untapped treasure trove" that the biomedical
research community could use to better understand diseases and their treatments.
"The automated de-identification software developed under the guidance of Dr.
Mark is a big step forward in permitting the widespread sharing of patient information
without the risk of compromised privacy and confidentiality," Cohen said.
Clifford, Mark and colleagues tested their censoring software on 1,836 nursing notes
(a total of 296,400 words). Using multiple experts and additional algorithms, they
replaced all personal information with "fake" data. In their BMC paper,
they report that "the software successfully deleted more than 94 percent of
the confidential information, while wrongly deleting only 0.2 percent of the useful
content. This is significantly better than one expert working alone, at least as
good as two trained medical professionals checking each other's work and many,
many times faster than either."
The team is providing researchers access to the evaluation dataset together with
the software to allow others to improve their systems, and to allow the software
to be adapted to other data types that may exhibit different qualities.
Clifford and Mark's co-authors are Ishna Neamatullah; Margaret M. Douglass;
Li-wei H. Lehman, an HST research engineer; Andrew Reisner, an HST visiting scientist;
Mauricio Villarroel, an HST visiting engineer; William J. Long, a principal research
associate in MIT's Computer Science and Artificial Intelligence Laboratory (CSAIL);
Professor Peter Szolovits of the Department of Electrical Engineering and Computer
Science and HST; and George B. Moody, HST sponsored research staff.
Back to Top
August
In Memoriam: Dr. Robert F. Wagner (1938-2008)
Dr. Robert F. Wagner was a distinguished research physicist and member of the Senior
Biomedical Research Service in the Center for Devices and Radiological Health (CDRH),
U.S. Food and Drug Administration (FDA). His career was dedicated to the development
of consensus measurement methods for the assessment of medical imaging systems,
quantitative medical imaging and tissue characterization, and computer-aided diagnosis.
He earned an international reputation in these areas and applied his expertise to
a wide range of regulatory issues central to FDA’s mission. He enlightened
the scientific community within the Agency as well as the international scientific
community through many invited presentations and tutorials, his numerous publications,
his many professional society activities, and his assistance in regulatory decision
making. His greatest legacy may be the many young scientists he nurtured, either
by direct tutelage at the FDA or by his many teaching engagements and research collaborations.
Robert F. Wagner was born in Philadelphia on January 10, 1938. After graduate and
post-graduate work on the physics of nuclear interactions with radiation, he was
hired by the Bureau of Radiological Health (a precursor to CDRH) to assess the dose
reduction potential of radiographic intensifying screens. Within months he published
a review of the relevant imaging literature together with a charter for a laboratory
program. Soon thereafter, Bob introduced digital noise analysis to radiography,
and showed that the new technology offered a 1.6 to 2.5-fold exposure reduction
without compromising imaging performance. He then launched a program of inter-laboratory
comparison of measurements on radiographic film samples that were circulated among
fifteen commercial, government, and academic laboratories world wide. In the process
he became the prime mover for work toward consensus methodology for quantitative
imaging performance measurements.
In the mid-1970s most investigators were concerned by the high exposures and doses
required by the newly introduced CT technology. Wagner and his colleagues showed
that CT was intrinsically a high-dose technique and his analyses and clarifications
were used by the BRH laboratory staff in the writing of the CT amendments to the
X-ray Performance Standards. Bob Wagner subsequently led the development of a “unified
signal-to-noise” ratio approach to the assessment of all conventional modes
of medical imaging. This work led to the wide use of detection quantum efficiency
as a performance metric for radiographic imaging systems that continues to this
day.
Dr. Wagner’s valuable ability to unify performance measures among different
imaging was next applied to ultrasound. Adopting key results from radar and laser
literature, he was able to develop the first satisfactory theoretical and experimental
approach to quantitative understanding of ultrasound speckle, its dependence on
ultrasonic hardware parameters, and its effect on the detectability of lesions by
medical ultrasound. Two award winning papers in 1983 and a stream of related publications
on ultrasound that followed made possible many advances in the development of standards
for ultrasonic imaging system performance assessment. In another prize winning 1989
Investigative Radiology publication, he showed that the incoherent backscattered
signal could be separated from two classes of coherent ultrasound signals returned
from the body and demonstrated how the resulting algorithm discriminated several
diffuse liver diseases from each other and from normal liver.
In the field of decision analysis, Dr. Wagner and his collaborators published key
investigations on the ability of human observers to extract information from images
obtained by a medical imaging system. In a 1981 article in Science, Dr. Wagner extended
his investigations of human performance to explore statistical methods for analyzing
the performance of imaging systems within the context of reader variability. Thereafter,
he extended his image assessment methodology to the many new and increasingly sophisticated
computer techniques being developed to aid the human reader in the interpretation
of high-dimensional image data sets for medical diagnosis. In recent years, he was
working on study designs, objective measurements, and analytical methods for the
assessment of stand-alone diagnostic modalities such as high-dimensional DNA micro-arrays.
Dr. Wagner’s research resulted in a stream of highly cited and extremely creative
scientific publications, as well as recognition in the form of honors and awards
by FDA and his professional community. He was elected to the rank of Fellow by five
societies: IEEE, AIMBE, OSA, SPIE, and SPSE. Within the FDA he was awarded the FDA
Commendable Service Award and the Award of Merit, the Commissioner’s Special
Citation, and the Public Health Service Superior Service award, in addition to having
been a member of numerous groups receiving unit awards. In recognition of his contributions,
Dr. Wagner was chosen as a principal author of an International Commission for Radiation
Units and Measurements (ICRU) report on image quality in medical imaging. This 1995
document laid the foundation for a series of ICRU reports with more detailed recipes,
one medical imaging modality at a time, that have been developed since. He served
on numerous academic advisory boards, search committees, conference program committees,
and editorial boards, and carried on an extremely active service function as a reviewer
of grants and scientific papers.
Dr. Wagner revolutionized the way in which medical imaging technologies are evaluated.
He spent more than 35 years in seminal work on understanding and developing paradigms
for the description and evaluation of medical imaging and computer-assisted diagnostic
modalities. His contributions to the science enterprise will live on in the work
of his students and colleagues for decades to come.
Back to Top
September
NIBIB Welcomes Two New Members to Advisory Council
Two new members were recently appointed to the Advisory Council of the National
Institute of Biomedical Imaging and Bioengineering (NIBIB). The Council serves as
the principal advisory body to the NIBIB, a component of the National Institutes
of Health. The Council, which meets three times a year, provides recommendations
on research priority and opportunities in biomedical imaging and bioengineering
and research training.
At the meeting on September 16, 2008, NIBIB Director Roderic I. Pettigrew, Ph.D.,
M.D., introduced the following new members:
PHILIP O. ALDERSON, M.D., is the Dean of the Saint Louis University School of Medicine,
a position he assumed in April 2008. He is a renowned nuclear medicine physician
and diagnostic radiologist who helped develop standard procedures for noninvasive
diagnosis of pulmonary emboli. Dr. Alderson is a Past President of the Academy for
Radiology Research. Prior to joining Saint Louis University, he was the chairman
of the department of radiology at Columbia-Presbyterian Medical Center and the James
Picker Professor of Radiology at the College of Physicians and Surgeons at Columbia
University. While at Columbia, he championed the integration of bioengineering and
radiology, and promoted the rapidly developing area of molecular imaging. Dr. Alderson
received his medical degree from Washington University in St. Louis.
CHERRI M. PANCAKE, PH.D., is a professor of electrical engineering and computer
science and Intel faculty fellow at Oregon State University. She is a pioneer in
applying ethnographic techniques to identify software usability problems of science
and business communities. The methods she developed are used in software products
from Hewlett Packard, Convex, Intel, IBM, and Tektronix. Recently, she has focused
on how virtual collaborations differ from proximal collaborations. Dr. Pancake received
her degree in computer engineering from Auburn University. Her research interests
are in usability engineering, more specifically, addressing the problem of how complex
software can better support the conceptual models and computing strategies of practicing
scientists and engineers. Dr. Pancake has been instrumental in the creation of the
Parallel Tools Consortium and the Network for Earthquake Engineering Simulation
(NEESH).
Members of the Advisory Council are drawn from the scientific communities, appointed
for 4-year terms, and represent all areas within the Institute's research mission.
Back to Top
April
April 4: Bioimaging's New Dynamic Duo: PET/MRI
Integration
Seeking to exploit the best features of two different bioimaging systems, researchers
in Dr. Simon Cherry’s molecular imaging lab at the University of California
Davis (UC Davis), are combining two very familiar technologies—positron emission
tomography (PET) and magnetic resonance imaging (MRI)—to create a more versatile
imaging tool. With collaborative contributions from Dr. Russell Jacob’s team
at the California Institute of Technology and Dr. Bernd Pichler of Tubingen University
in Germany, results of this tandem PET/MRI research, which is sponsored by NIBIB,
indicate this new technology could lead to improvements in many aspects of health
care, as well as in basic research.
Both imaging systems have unique strengths that compliment each other. For example,
PET is much more sensitive than MRI. Because it can zero in on very faint signals
emitted by radioactively labeled sugar molecules that enter cancer cells, PET is
often used to locate tumors. On the other hand, one of MRI’s strengths is
its ability to produce clearly defined images of anatomical structures and soft
tissues, which can provide critical information such as changes in the size of a
tumor following treatment.
This team imaging approach has potential to help new treatments get from the laboratory
to the patient’s point of care in less time. Dr. Cherry explains, “These
two technologies are powerful allies in the evaluation of new medical therapies
in animal models where the detailed information they provide on the distribution
and action of a drug can help improve the success of the translation of new therapies
into the clinic.”
Today, many physicians use data from these two imaging modalities to evaluate anatomy,
physiology, cellular metabolism, molecular targets and pathways, and even gene expression.
This information helps build a strong foundation of evidence for biological or medical
evaluation that helps doctors determine, and monitor, a medical course of action.
Moreover, these systems allow clinicians to peer deep into the body without performing
surgery or other invasive techniques.
With all this in mind, one might ask why researchers have not previously combined
these modalities, especially given the success of other combined imaging systems
such as PET and x-ray computed tomography (CT). The answer to that question lies
in the many ways that the two systems can interfere with each other and distort
the images that are recorded. For example the detectors used in most PET scanners
are extremely sensitive to magnetic fields and will not operate inside the strong
magnetic field of an MRI scanner. Furthermore, in order to obtain a clear MRI image,
its magnetic field must be stable and uniform, but the mere placement of PET detectors
within the scanner can disrupt the magnetic field.
For these reasons, PET and MRI scans have previously been done independently, but
the problem in using these imaging systems independently is that the process of
accurately weaving both sets of information together can be very difficult. The
process works fairly well in applications such as brain imaging where there is little
potential for movement or change in position, but the true challenge arises in locations
such as the chest or abdomen where movement can be problematic, and if MRI and PET
data cannot be matched or “registered” perfectly, interpretation of
images can be difficult. Furthermore, much important biological information would
be revealed about dynamic processes if we had the ability to observe such events
using both systems at the same time. Therefore, development of a dual system capable
of providing simultaneous PET and MRI scans is a great benefit in situations where
movement is inevitable, and it also provides the added benefit of real-time, dual-imaging
observations of physiological processes.
Realizing the tremendous benefits of such a system, researchers at UC Davis have
designed a new integrated PET/MRI scanner that positions a PET scanner within the
MRI scanner, while also minimizing the interference between the two systems by employing
special materials and clever design strategies. For example, the new integrated
PET system’s data is decoded using special solid-state detectors (developed
by Radiation Monitoring Devices, Inc., with NIH and Department of Energy Small Business
Innovation Research funding) that can tolerate the strong magnetic fields inside
the MRI. Detectors are also shielded by a high-frequency copper laminate and kept
at a constant, cool negative 10 degrees Celsius (14 degrees Fahrenheit) to optimize
results. Additionally, the short optical fiber bundles are positioned precisely
to further minimize interference. A convenient feature of the new system is that
the PET scanner insert fits many commercially available MRI systems and can be easily
removed from the MRI scanner so that the units can be used separately, if necessary.
Initial studies have clearly demonstrated the promise of the PET/MRI technology,
and Dr. Cherry predicts, “While the technology will continue to improve and
mature, the spotlight will now shift to finding unique ways to use this integrated
technology to learn new things about the biology of disease and the best way to
use PET/MRI for early diagnosis and to monitor treatment response.”
Researchers already predict that the new system has the potential to greatly enhance
our understanding of the origins and evolution of disease processes, such as cancer,
Alzheimers, and bone disease. It will also help monitor the effectiveness of cancer
treatments, locate metastases in soft tissues, and give clues about tumor physiology
and the immune system’s ability to fight cancer.
Dual-modality imaging can yield data that automatically match up in time and space
because both PET and MRI imaging scans of the same body regions are done at exactly
the same time. Even in brain tissues where independent use of these technologies
has been very useful, the new system can do more, such as PET analysis of glucose
metabolism while MRI images evaluate constriction or dilation of blood vessels.
As for nurturing this fledgling technology to its full potential, this collaborative
research team is already working on it. The integrated PET/MRI system is now being
used for preclinical research, and the UC Davis team is developing ways to further
improve on their prototype technology by increasing the spatial resolution and sensitivity
of the PET component.
So even just as the nuptial ceremonies are coming to a close and the union of these
imaging systems is sealed, investigators are brainstorming about the many possibilities
for its future. And just like Dr. Charles Townes’s famous invention, the laser,
the full potential of PET/MRI integration will only be realized as its offspring
emerge from the pipeline of this new wave of technology, arriving as creative new
technologies in their own right in the healthcare of our future.
If you would like to learn more about Genomic Imaging research at UC Davis, see
Dr. Cherry's web site at:
http://www.bme.ucdavis.edu/profiles/cherry.html ,
or see, "Simultaneous in vivo positron emission tomography and magnetic
resonance imaging" at:
http://www.pnas.org/cgi/content/abstract/105/10/3705
,
or see, "Simultaneous in vivo positron emission tomography and magnetic
resonance imaging" at:
http://www.pnas.org/cgi/content/abstract/105/10/3705 .
.
Back to Top
June
June 18: NIST/NIH Micromagnets Show Promise as Colorful "Smart Tags" for
Magnetic Resonance Imaging
BOULDER, Colo.—Customized microscopic magnets that might one day be injected
into the body could add color to magnetic resonance imaging (MRI), while also potentially
enhancing sensitivity and the amount of information provided by images, researchers
at the National Institute of Standards and Technology (NIST) and National Institutes
of Health (NIH) report. The new micromagnets also could act as “smart tags”
identifying particular cells, tissues, or physiological conditions, for medical
research or diagnostic purposes.
|

Microscopic magnets (above left), designed and tested in a joint NIST/NIH project,
might one day be injected into the body to add color and “smart tag”
capability to magnetic resonance imaging for medical diagnosis and research. The
image on the right shows light scattering from grids of magnets on a wafer where
they were made using conventional microfabrication techniques.
Credit: G. Zabow, NIST/NIH
View hi-resolution of image on left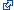 View hi-resolution of image on right
View hi-resolution of image on right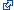 |
|
As described in the June 19 issue of Nature,* the NIST and NIH investigators have
demonstrated the proof of principle for a new approach to MRI. Unlike the chemical
solutions now used as image-enhancing contrast agents in MRI, the NIST/NIH micro-magnets
rely on a precisely tunable feature—their physical shape—to adjust the
radio-frequency (RF) signals used to create images. The RF signals then can be converted
into a rainbow of optical colors by computer. Sets of different magnets designed
to appear as different colors could, for example, be coated to attach to different
cell types, such as cancerous versus normal. The cells then could be identified
by tag color.
“Current MRI technology is primarily black and white. This is like a colored
tag for MRI,” says lead author Gary Zabow, who designed and fabricated the
microtags at NIST and, together with colleagues at the National Institute of Neurological
Disorders and Stroke, part of NIH, tested them on MRI machines.
The micromagnets also can be thought of as microscopic RF identification (RFID)
tags, similar to those used for identifying and tracking objects from nationwide
box shipments to food in the supermarket. The device concept is flexible and could
have other applications such as in enabling RFID-based microscopic fluid devices
(microfluidics) for biotechnology and handheld medical diagnostic toolkits.
The microtags would need extensive further engineering and testing, including clinical
studies, before they could be used in people undergoing MRI exams. The magnets used
in the NIST/NIH studies were made of nickel, which is toxic but was relatively easy
to work with for the initial prototypes. But Zabow says they could be made of other
magnetic materials, such as iron, which is considered non-toxic and is already approved
for use in certain medical agents. Only very low concentrations of the magnets would
be needed in the body to enhance MRI images.
Each micromagnet consists of two round, vertically stacked magnetic discs a few
micrometers in diameter, separated by a small open gap in between. Researchers create
a customized magnetic field for each tag by making it from particular materials
and tweaking the geometry, perhaps by widening the gap between the discs or changing
the discs’ thickness or diameter. As water in a sample flows between the discs,
protons acting like twirling bar magnets within the water’s hydrogen atoms
generate predictable RF signals—the stronger the magnetic field, the faster
the twirling—and these signals are used to create images.
The open sandwich design allows the movement or diffusion of water through the micromagnet,
producing a signal that may be thousands of times stronger than that produced by
a similarly sized, but stationary, volume of water. The diffusion effectively increases
local MRI sensitivity, which in a future clinical setting could lead to practical
benefits such as faster imaging, images that are richer with information, or reduced
dose requirements for these contrast agents. The NIST/NIH test results show that
changing magnet geometry results in significant shifts in the frequency signals.
Thanks to their physical attributes, the magnets can be designed to have more tunable
properties than conventional injectable MRI contrast agents. MRI contrast agents
enhance images by altering the magnetic field seen by hydrogen nuclei in water.
Conventional contrast agents are chemically synthesized whereas the new micromagnets
are microfabricated. This allows for greater control and range of the modified magnetic
field, greatly enhancing sensitivity.
Furthermore, unlike the molecular chemical “soups” that make up many
of the contrast agents, each micromagnet potentially could be individually detected
for imaging purposes. The magnets also could be designed to be turned on and off
by, for example, filling the gap between the discs to block water passage. The gap
could be filled with something that dissolves when exposed to certain substances
or conditions, Zabow says.
The micromagnets can be made using conventional microfabrication techniques and
are compatible with standard MRI hardware. Advanced lithography techniques of the
kind used to make sophisticated computer chips might be used to make the tags even
smaller, approaching the nanometer scale, according to the paper.
The magnets could make medical diagnostic images as information-rich as the optical
images of tissue samples now common in biotechnology, which already benefits from
a variety of colored markers such as fluorescent proteins and tunable quantum dots.
NIH has filed a provisional patent application on the micromagnets.
NIH support for Zabow’s work was funded by the National Institute of Biomedical
Imaging and Bioengineering (NIBIB) through the NIST/NIH-NIBIB National Research
Council Joint Associateship Program. The program seeks to recruit physicists into
biomedical research in order to improve technologies like MRI.
As a non-regulatory agency, NIST promotes U.S. innovation and industrial competitiveness
by advancing measurement science, standards and technology in ways that enhance
economic security and improve our quality of life.
The National Institutes of Health (NIH)—The Nation's Medical Research
Agency—includes 27 institutes and centers and is a component of the U.S. Department
of Health and Human Services. It is the primary federal agency for conducting and
supporting basic, clinical and translational medical research, and it investigates
the causes, treatments and cures for both common and rare diseases. For more information
about NIH and its programs, visit www.nih.gov.
*G. Zabow, S. Dodd, J. Moreland, A. Koretsky. 2008. Micro-engineered local field
control for high-sensitivity multispectral MRI. Nature. June 19.
Back to Top
Last Updated On 10/18/2011