A New Method to Help Scientists Better Identify Drug Candidates
In the past few years, researchers have discovered that the use of reporter genes, a powerful technique widely used in drug discovery screening, can produce misleading results and lead to wasted effort and inefficiency in the drug discovery process. Now, researchers from NIH’s National Center for Advancing Translational Sciences (NCATS) have designed a novel method that increases the odds of identifying candidate compounds with true activity against biological or disease targets. Scientists use these compounds as molecular tools to understand disease and as starting points for developing new therapeutics. Details of the methodology are described in a correspondence published in the October issue of Nature Methods.
A Powerful Technique
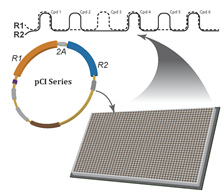
The two reporter-gene solution (depicted above as R1 and R2 measured simultaneously and independently in the assay plate) is based on the principle that it is easier to distinguish signal from ‘noise’ when the signal noting biological activity is reported by two or more detectors.
Reporter genes produce proteins that act as light-generating (e.g., luminescent or fluorescent) sentinels when a chemical compound produces an effect in a testing system (an "assay") designed to represent a biological or disease process. Such assays enable researchers to examine hundreds of thousands of compounds using robotic high-throughput screening systems, such as those in the NCATS Division of Pre-Clinical Innovation (DPI). Typically, scientists use a single reporter gene for screening; however, because chemical compounds may interact with the reporter gene proteins themselves, rather than with the intended target, these tests can be misleading.
Compounds affecting the reporter may inhibit its activity, or alternatively may bind to and stabilize it, increasing its abundance and activity in the cell. In either case, a researcher may see a strong effect of the compound in the assay, and erroneously conclude that this indicates action on the biological pathway or target of interest. But, it is more a case of the reporter doing "false advertising," leading the researcher to conduct further testing and development, and even clinical trials, that ultimately fail since the compound only has activity on the reporter, not the disease target.
"The mission of NCATS is to develop, demonstrate, and disseminate technologies and paradigms that improve the accuracy, efficiency and predictability of the translational process," said Christopher P. Austin, M.D., NCATS director. "This innovative reporter gene system is a great example of such a platform technology, which will enable researchers worldwide in achieving more reliable results early in the translational process, and avoiding failure at later stages."
A Method Made Better
The new method uses two co-expressed reporter genes rather than one. DPI researcher James Inglese, Ph.D., and his postdoctoral fellow Ken C-C Cheng, Ph.D., designed the new method, calling it a "coincidence reporter-gene system for high-throughput screening." When contemplating the creation of the method, they knew they could not control compounds from interacting with reporters or predict in what situation it may occur. To overcome this, the new method uses a short DNA sequence specially designed to allow the equal expression and proper function of two mechanistically unrelated reporters in the same cell to search for coinciding signals in high-throughput drug screens. For academic researchers who rely on reporter-genes, adding an additional reporter is easy to do, and the plasmids to enable this new method are available through the not-for-profit plasmid repository, Addgene.
The two reporter-gene solution is based on the principle that it is easier to distinguish signal from "noise" when the signal is reported by two or more detectors. Such coincidence detectors are a strategy employed in other fields. For instance, Positron Emission Tomography (PET) scans use this technology to detect cancerous tumors. In astrophysics, they are used to detect signals from space. And in the human body, our brain uses coincidence detection as part of its neural network.
Building on this theory, Inglese and Cheng thought that if the reporter genes could be used together, researchers might be able to see instances when both reporters were active, providing more confidence that the test compound was having the sought after effect. To test this, they used firefly luciferase (FLuc) and Renilla luciferase (RLuc). As a result, the researchers were able to demonstrate the accuracy of the method by conducting a high-throughput screen to discriminate compounds with activity on a well-studied biological pathway versus those that were artifacts of direct reporter-gene interaction.
"We now can use this method to see what signals are common between the two distinct reporters as an indication of what molecules should be pursued further," said Inglese, head of the Assay Development and Screening Technology Laboratory in DPI. "The presence of two reporter-gene signals translates to a higher probability of a biological response rather than a compound interacting with a reporter."
According to Inglese, the improved method will save researchers a great deal of time and money in the long run. The next steps for his laboratory are to understand the increase in efficiency gained from using the new methodology, and whether multiple multiple reporter-gene combinations can further extend the fidelity and sensitivity of a response.
"Our method represents a simple and accessible solution to a complex and pervasive problem," Inglese said. "We are confident that scientists can use it to increase the predictability of drug screening results."
Posted November 2012
Social Media Links