Reviewed February 2012
What is chromosome 4?
Humans normally have 46 chromosomes in each cell, divided into 23 pairs. Two copies of chromosome 4, one copy inherited from each parent, form one of the pairs. Chromosome 4 spans more than 191 million DNA building blocks (base pairs) and represents more than 6 percent of the total DNA in cells.
Identifying genes on each chromosome is an active area of genetic research. Because researchers use different approaches to predict the number of genes on each chromosome, the estimated number of genes varies. Chromosome 4 likely contains between 1,300 and 1,600 genes. These genes perform a variety of different roles in the body.
Genes on chromosome 4 are among the estimated 20,000 to 25,000 total genes in the human genome.
Genetics Home Reference includes these genes on chromosome 4:
-
AGA
-
ANK2
-
ANTXR2
-
CFI
-
CISD2
-
CYP4V2
-
DOK7
-
DRD5
-
DSPP
-
ENAM
-
ETFDH
-
EVC
-
EVC2
-
FGA
-
FGB
-
FGFR3
-
FGFRL1
-
FGG
-
FIP1L1
-
HADH
-
HTT
-
IDUA
-
KIT
-
LETM1
-
MANBA
-
MMAA
-
MSX1
-
MTTP
-
NR3C2
-
PDGFRA
-
PHOX2B
-
PITX2
-
PKD2
-
QDPR
-
SGCB
-
SH3BP2
-
SLC25A4
-
SNCA
-
TET2
-
UCHL1
-
UVSSA
-
WFS1
-
WHSC1
How are changes in chromosome 4 related to health conditions?
Many genetic conditions are related to changes in particular genes on chromosome 4.
This list of disorders associated with genes on chromosome 4 provides links to additional information.
Genetics Home Reference includes these conditions related to genes on chromosome 4:
- abetalipoproteinemia
- achondroplasia
- age-related macular degeneration
- amelogenesis imperfecta
- aspartylglucosaminuria
- atypical hemolytic-uremic syndrome
- Axenfeld-Rieger syndrome
- benign essential blepharospasm
- beta-mannosidosis
- Bietti crystalline dystrophy
- bladder cancer
- cherubism
- complement factor I deficiency
- congenital afibrinogenemia
- congenital central hypoventilation syndrome
- congenital myasthenic syndrome
- Crouzonodermoskeletal syndrome
- dentinogenesis imperfecta
- Ellis-van Creveld syndrome
- epidermal nevus
- essential thrombocythemia
- familial hyperinsulinism
- gastrointestinal stromal tumor
- glutaric acidemia type II
- Huntington disease
- 3-hydroxyacyl-CoA dehydrogenase deficiency
- hypochondroplasia
- infantile systemic hyalinosis
- juvenile hyaline fibromatosis
- limb-girdle muscular dystrophy
- methylmalonic acidemia
- mucopolysaccharidosis type I
- Muenke syndrome
- multiple system atrophy
- neuroblastoma
- nonsyndromic deafness
- Parkinson disease
- PDGFRA-associated chronic eosinophilic leukemia
- piebaldism
- polycystic kidney disease
- polycythemia vera
- primary myelofibrosis
- progressive external ophthalmoplegia
- pseudohypoaldosteronism type 1
- Romano-Ward syndrome
- SADDAN
- tetrahydrobiopterin deficiency
- thanatophoric dysplasia
- UV-sensitive syndrome
- Weyers acrofacial dysostosis
- Wolf-Hirschhorn syndrome
- Wolfram syndrome
Changes in the structure or number of copies of a chromosome can also cause problems with health and development. The following chromosomal conditions are associated with such changes in chromosome 4.
- cancers
-
Changes in chromosome 4 have been identified in several types of human cancer. These genetic changes are somatic, which means they are acquired during a person's lifetime and are present only in certain cells. For example, rearrangements (translocations) of genetic material between chromosome 4 and several other chromosomes have been associated with leukemias, which are cancers of blood-forming cells.
A specific translocation involving chromosome 4 and chromosome 14 is commonly found in multiple myeloma, which is a cancer that starts in cells of the bone marrow. The translocation, which is written as t(4;14)(p16;q32), abnormally fuses the WHSC1 gene on chromosome 4 with part of another gene on chromosome 14. The fusion of these genes overactivates WHSC1, which appears to promote the uncontrolled growth and division of cancer cells.
- facioscapulohumeral muscular dystrophy
-
Facioscapulohumeral muscular dystrophy results from a deletion of genetic material from a region of DNA known as D4Z4. This region is located near the end of the long (q) arm of chromosome 4 at a position described as 4q35. The D4Z4 region normally consists of 11 to more than 100 repeated DNA segments, each of which is about 3,300 DNA base pairs (3.3 kb) long. However, in people with facioscapulohumeral muscular dystrophy the D4Z4 region on one copy of chromosome 4 is abnormally short, containing between 1 and 10 repeats.
It is uncertain how a shortened D4Z4 region causes the progressive muscle weakness and wasting characteristic of facioscapulohumeral muscular dystrophy. Researchers have proposed several possible mechanisms, but none have yet been proven. It appears that the D4Z4 region influences the activity of other genes located nearby on the long arm of chromosome 4. An abnormally short D4Z4 region may somehow disrupt the normal regulation of these genes. However, it is unclear which genes are influenced by D4Z4 and what role, if any, those genes play in muscle cells. Researchers suspect that genetic factors other than the shortened D4Z4 region may also be involved in facioscapulohumeral muscular dystrophy.
- PDGFRA-associated chronic eosinophilic leukemia
-
PDGFRA-associated chronic eosinophilic leukemia is caused by genetic abnormalities that involve the PDGFRA gene, a gene found on chromosome 4. This condition is a type of blood cell cancer characterized by an increased number of eosinophils, a type of white blood cell involved in allergic reactions.
The PDGFRA gene abnormalities are somatic mutations, which are mutations acquired during a person's lifetime that are present only in certain cells. The most common of these abnormalities is a deletion of genetic material from chromosome 4 that removes approximately 800 DNA building blocks (nucleotides) and brings together parts of two genes, FIP1L1 and PDGFRA, creating the FIP1L1-PDGFRA fusion gene. Occasionally, through mechanisms other than deletion, genes other than FIP1L1 are fused with the PDGFRA gene. Rarely, mutations that change single DNA building blocks in the PDGFRA gene (point mutations) cause this condition.
The protein produced from the FIP1L1-PDGFRA fusion gene (as well as other PDGFRA fusion genes) has the function of the PDGFRA protein, which stimulates signaling pathways inside the cell that control many important cellular processes, such as cell growth and division (proliferation) and cell survival. Unlike the normal PDGFRA protein, however, the fusion protein is constantly turned on (constitutively activated), which means the cells are always receiving signals to proliferate. Similarly, point mutations in the PDGFRA gene can result in a constitutively activated PDGFRA protein. When the FIP1L1-PDGFRA fusion gene or point mutations in the PDGFRA gene occur in blood cell precursors, the growth of eosinophils (and occasionally other blood cells) is poorly controlled, leading to PDGFRA-associated chronic eosinophilic leukemia. It is unclear why eosinophils are preferentially affected by this genetic change.
- Wolf-Hirschhorn syndrome
-
Wolf-Hirschhorn syndrome is caused by a deletion of genetic material near the end of the short (p) arm of chromosome 4 at a position described as 4p16.3. The signs and symptoms of this condition are related to the loss of multiple genes from this part of the chromosome. The size of the deletion varies among affected individuals; studies suggest that larger deletions tend to result in more severe intellectual disability and physical abnormalities than smaller deletions.
The region of chromosome 4 that is deleted most often in people with Wolf-Hirschhorn syndrome is known as Wolf-Hirschhorn syndrome critical region 2 (WHSCR-2). This region contains several genes, some of which are known to play important roles in early development. A loss of these genes leads to developmental delay, a distinctive facial appearance, and other characteristic features of the condition. Scientists are working to identify additional genes at the end of the short arm of chromosome 4 that contribute to the characteristic features of Wolf-Hirschhorn syndrome.
- other chromosomal conditions
-
Some deletions of genetic material from the short (p) arm of chromosome 4 do not involve the critical region WHSCR-2. These deletions cause signs and symptoms that are distinct from those of Wolf-Hirschhorn syndrome, including mild intellectual disability and, in some cases, rapid (accelerated) growth. People with this type of deletion usually do not have seizures.
Trisomy 4 occurs when cells have three copies of chromosome 4 instead of the usual two copies. Full trisomy 4, which occurs when all of the body's cells contain an extra copy of chromosome 4, is not compatible with life. A similar but somewhat less severe condition called mosaic trisomy 4 occurs when only some of the body's cells have an extra copy of chromosome 4. The signs and symptoms of mosaic trisomy 4 vary widely and can include heart defects, abnormalities of the fingers and toes, and other birth defects. Mosaic trisomy 4 is very rare; only a few cases have been reported.
Other changes in the number or structure of chromosome 4 can have a variety of effects including delayed growth and development, intellectual disability, distinctive facial features, heart defects, and other medical problems. Changes involving chromosome 4 include an extra piece of the chromosome in each cell (partial trisomy 4), a missing segment of the chromosome in each cell (partial monosomy 4), and a circular structure called a ring chromosome 4. Ring chromosomes occur when a chromosome breaks in two places and the ends of the chromosome arms fuse together to form a circular structure.
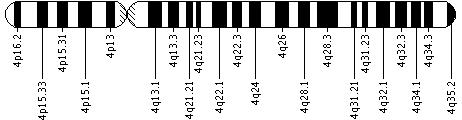
Is there a standard way to diagram chromosome 4?
Geneticists use diagrams called ideograms as a standard representation for chromosomes. Ideograms show a chromosome's relative size and its banding pattern. A banding pattern is the characteristic pattern of dark and light bands that appears when a chromosome is stained with a chemical solution and then viewed under a microscope. These bands are used to describe the location of genes on each chromosome.
See How do geneticists indicate the location of a gene? (http://ghr.nlm.nih.gov/handbook/howgeneswork/genelocation) in the Handbook.
Where can I find additional information about chromosome 4?
You may find the following resources about chromosome 4 helpful. These materials are written for the general public.
You may also be interested in these resources, which are designed for genetics professionals and researchers.
-
Gene Reviews - Clinical summary
- Gene Review: Facioscapulohumeral Muscular Dystrophy (http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=gene&part=fsh)
- Gene Review: Wolf-Hirschhorn Syndrome (http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=gene&part=whs)
-
Gene Tests - Genetic tests ordered by healthcare professionals
- Gene Tests: Facioscapulohumeral Muscular Dystrophy (http://www.ncbi.nlm.nih.gov/sites/GeneTests/lab/clinical_disease_id/2640)
- Gene Tests: Wolf-Hirschhorn Syndrome (http://www.ncbi.nlm.nih.gov/sites/GeneTests/lab/clinical_disease_id/2594)
-
Research Resources - Tools for researchers
- Atlas of Genetics and Cytogenetics in Oncology and Haematology: Chromosome 4 (http://atlasgeneticsoncology.org/Indexbychrom/idxa_4.html)
- Atlas of Genetics and Cytogenetics in Oncology and Haematology: t(4;14)(p16;q32) (http://atlasgeneticsoncology.org/Anomalies/t04142059.html)
- Cancer Genetics Web (http://www.cancerindex.org/geneweb/clinkc04.htm)
- Database of Genomic Variants (http://projects.tcag.ca/variation/cgi-bin/tbrowse/tbrowse?source=hg17&table=Locus&show=table&keyword=&flop=AND&fcol=_C19&fcomp==&fkwd=chr4&cols=)
- Ensembl Human Map View (http://www.ensembl.org/Homo_sapiens/Location/Chromosome?chr=4)
- Generation and annotation of the DNA sequences of human chromosomes 2 and 4. Nature. 2005 Apr 7;434(7034):724-31. (http://www.nature.com/nature/journal/v434/n7034/full/nature03466.html)
- U.S. Department of Energy: Human Chromosome Launchpad (http://www.ornl.gov/sci/techresources/Human_Genome/launchpad/chrom04.shtml)
- PubMed - Recent literature (http://www.ncbi.nlm.nih.gov/pubmed?term=(Chromosomes,%20Human,%20Pair%204%5BMAJR%5D)%20AND%20((4%5BTI%5D)%20OR%20(4p%5BTI%5D)%20OR%20(4q%5BTI%5D))%20AND%20english%5Bla%5D%20AND%20human%5Bmh%5D%20AND%20%22last%20720%20days%22%5Bdp%5D)
- OMIM - Genetic disorder catalog (http://omim.org/entry/254500)
- Map Viewer - Genetic maps (http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?org=human&maps=ideogr,morbid,pheno&zoom=100&chr=4)
What glossary definitions help with understanding chromosome 4?
base pair ;
birth defect ;
bone marrow ;
cancer ;
cell ;
chromosome ;
chronic ;
critical region ;
deletion ;
developmental delay ;
DNA ;
DNA base ;
eosinophils ;
gene ;
kb ;
leukemia ;
monosomy ;
mosaic ;
multiple myeloma ;
muscular dystrophy ;
mutation ;
myeloma ;
nucleotide ;
point mutation ;
proliferate ;
proliferation ;
protein ;
rearrangement ;
ring chromosomes ;
seizure ;
sign ;
somatic mutation ;
symptom ;
syndrome ;
translocation ;
trisomy ;
wasting ;
white blood cells
You may find definitions for these and many other terms in the Genetics Home Reference
Glossary (http://ghr.nlm.nih.gov/glossary).
References
- Bergemann AD, Cole F, Hirschhorn K. The etiology of Wolf-Hirschhorn syndrome. Trends Genet. 2005 Mar;21(3):188-95. Review. (http://www.ncbi.nlm.nih.gov/pubmed/15734578?dopt=Abstract)
- Buitenhuis M, Verhagen LP, Cools J, Coffer PJ. Molecular mechanisms underlying FIP1L1-PDGFRA-mediated myeloproliferation. Cancer Res. 2007 Apr 15;67(8):3759-66. (http://www.ncbi.nlm.nih.gov/pubmed/17440089?dopt=Abstract)
- Chen CP, Chern SR, Lee CC, Chang TY, Wang W, Tzen CY. Clinical, cytogenetic, and molecular findings of prenatally diagnosed mosaic trisomy 4. Prenat Diagn. 2004 Jan;24(1):38-44. Review. (http://www.ncbi.nlm.nih.gov/pubmed/14755408?dopt=Abstract)
- Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, Kutok J, Clark J, Galinsky I, Griffin JD, Cross NC, Tefferi A, Malone J, Alam R, Schrier SL, Schmid J, Rose M, Vandenberghe P, Verhoef G, Boogaerts M, Wlodarska I, Kantarjian H, Marynen P, Coutre SE, Stone R, Gilliland DG. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med. 2003 Mar 27;348(13):1201-14. (http://www.ncbi.nlm.nih.gov/pubmed/12660384?dopt=Abstract)
- Ensembl Human Map View (http://www.ensembl.org/Homo_sapiens/Location/Chromosome?chr=4)
- Goldfrank D, Schoenberger E, Gilbert F. Disease genes and chromosomes: disease maps of the human genome. Chromosome 4. Genet Test. 2003 Winter;7(4):351-72. (http://www.ncbi.nlm.nih.gov/pubmed/15000816?dopt=Abstract)
- Hayne CC, Winer E, Williams T, Chaves F, Khorsand J, Mark HF. Acute lymphoblastic leukemia with 4;11 translocation analyzed by a multi-modal strategy of conventional cytogenetics, FISH, morphology, flow cytometry and molecular genetics, and review of the literature. Exp Mol Pathol. 2006 Aug;81(1):62-71. Epub 2006 Jun 12. Review. (http://www.ncbi.nlm.nih.gov/pubmed/16765346?dopt=Abstract)
- Hillier LW, Graves TA, Fulton RS, Fulton LA, Pepin KH, Minx P, Wagner-McPherson C, Layman D, Wylie K, Sekhon M, Becker MC, Fewell GA, Delehaunty KD, Miner TL, Nash WE, Kremitzki C, Oddy L, Du H, Sun H, Bradshaw-Cordum H, Ali J, Carter J, Cordes M, Harris A, Isak A, van Brunt A, Nguyen C, Du F, Courtney L, Kalicki J, Ozersky P, Abbott S, Armstrong J, Belter EA, Caruso L, Cedroni M, Cotton M, Davidson T, Desai A, Elliott G, Erb T, Fronick C, Gaige T, Haakenson W, Haglund K, Holmes A, Harkins R, Kim K, Kruchowski SS, Strong CM, Grewal N, Goyea E, Hou S, Levy A, Martinka S, Mead K, McLellan MD, Meyer R, Randall-Maher J, Tomlinson C, Dauphin-Kohlberg S, Kozlowicz-Reilly A, Shah N, Swearengen-Shahid S, Snider J, Strong JT, Thompson J, Yoakum M, Leonard S, Pearman C, Trani L, Radionenko M, Waligorski JE, Wang C, Rock SM, Tin-Wollam AM, Maupin R, Latreille P, Wendl MC, Yang SP, Pohl C, Wallis JW, Spieth J, Bieri TA, Berkowicz N, Nelson JO, Osborne J, Ding L, Meyer R, Sabo A, Shotland Y, Sinha P, Wohldmann PE, Cook LL, Hickenbotham MT, Eldred J, Williams D, Jones TA, She X, Ciccarelli FD, Izaurralde E, Taylor J, Schmutz J, Myers RM, Cox DR, Huang X, McPherson JD, Mardis ER, Clifton SW, Warren WC, Chinwalla AT, Eddy SR, Marra MA, Ovcharenko I, Furey TS, Miller W, Eichler EE, Bork P, Suyama M, Torrents D, Waterston RH, Wilson RK. Generation and annotation of the DNA sequences of human chromosomes 2 and 4. Nature. 2005 Apr 7;434(7034):724-31. (http://www.ncbi.nlm.nih.gov/pubmed/15815621?dopt=Abstract)
- Keats JJ, Maxwell CA, Taylor BJ, Hendzel MJ, Chesi M, Bergsagel PL, Larratt LM, Mant MJ, Reiman T, Belch AR, Pilarski LM. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood. 2005 May 15;105(10):4060-9. Epub 2005 Jan 27. (http://www.ncbi.nlm.nih.gov/pubmed/15677557?dopt=Abstract)
- Keats JJ, Reiman T, Belch AR, Pilarski LM. Ten years and counting: so what do we know about t(4;14)(p16;q32) multiple myeloma. Leuk Lymphoma. 2006 Nov;47(11):2289-300. Review. (http://www.ncbi.nlm.nih.gov/pubmed/17107900?dopt=Abstract)
- Lundin C, Zech L, Sjörs K, Wadelius C, Annerén G. Trisomy 4q syndrome: presentation of a new case and review of the literature. Ann Genet. 2002 Apr-Jun;45(2):53-7. Review. (http://www.ncbi.nlm.nih.gov/pubmed/12119211?dopt=Abstract)
- Map Viewer: Genes on Sequence (http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?ORG=human&MAPS=ideogr,ugHs,genes&CHR=4)
- Pai, G Shashidhar; Lewandowski, Raymond C; Borgaonkar, Digamber S; Handbook of chromosomal syndromes; Hoboken, N.J. : Wiley-Liss, c2003.
- Roufosse FE, Goldman M, Cogan E. Hypereosinophilic syndromes. Orphanet J Rare Dis. 2007 Sep 11;2:37. Review. (http://www.ncbi.nlm.nih.gov/pubmed/17848188?dopt=Abstract)
- South ST, Hannes F, Fisch GS, Vermeesch JR, Zollino M. Pathogenic significance of deletions distal to the currently described Wolf-Hirschhorn syndrome critical regions on 4p16.3. Am J Med Genet C Semin Med Genet. 2008 Nov 15;148C(4):270-4. doi: 10.1002/ajmg.c.30188. (http://www.ncbi.nlm.nih.gov/pubmed/18932125?dopt=Abstract)
- Tawil R. Facioscapulohumeral muscular dystrophy. Neurotherapeutics. 2008 Oct;5(4):601-6. doi: 10.1016/j.nurt.2008.07.005. Review. (http://www.ncbi.nlm.nih.gov/pubmed/19019312?dopt=Abstract)
- UCSC Genome Browser: Statistics (http://genome.cse.ucsc.edu/goldenPath/stats.html)
- van der Maarel SM, Frants RR, Padberg GW. Facioscapulohumeral muscular dystrophy. Biochim Biophys Acta. 2007 Feb;1772(2):186-94. Epub 2006 Jun 6. Review. (http://www.ncbi.nlm.nih.gov/pubmed/16837171?dopt=Abstract)
- Zollino M, Lecce R, Fischetto R, Murdolo M, Faravelli F, Selicorni A, Buttè C, Memo L, Capovilla G, Neri G. Mapping the Wolf-Hirschhorn syndrome phenotype outside the currently accepted WHS critical region and defining a new critical region, WHSCR-2. Am J Hum Genet. 2003 Mar;72(3):590-7. Epub 2003 Jan 30. (http://www.ncbi.nlm.nih.gov/pubmed/12563561?dopt=Abstract)
- Zollino M, Murdolo M, Marangi G, Pecile V, Galasso C, Mazzanti L, Neri G. On the nosology and pathogenesis of Wolf-Hirschhorn syndrome: genotype-phenotype correlation analysis of 80 patients and literature review. Am J Med Genet C Semin Med Genet. 2008 Nov 15;148C(4):257-69. doi: 10.1002/ajmg.c.30190. Review. (http://www.ncbi.nlm.nih.gov/pubmed/18932124?dopt=Abstract)
The resources on this site should not be used as a substitute for
professional medical care or advice. Users seeking information about
a personal genetic disease, syndrome, or condition should consult with a qualified
healthcare professional.
See How can I find a genetics professional in my area? (http://ghr.nlm.nih.gov/handbook/consult/findingprofessional) in the Handbook.