Retinal Circuit Development & Genetics Unit
Building 6, Room 331
6 Center Drive
Bethesda, Maryland 20892- 0610
Research Goals
We are using molecular genetic approaches to study the development of neuronal cell types, with a particular focus on neuronal arbor formation in retinal neurons. Using sparse Cre-loxP recombination, we specifically label individual neurons or neuronal cell classes, and visualize their morphology and connectivity. By combining conditional gene ablation with reporter gene replacement, we are able to visualize neurons in which specific genes have been ablated, and study the consequences of loss of gene function on the development of the neurons. Targeting specific neuronal populations through mouse genetic manipulations enables us also to study the roles of these neurons in visual circuitry.
Current research directions in the lab:
Developing methodologies for sparse genetic labeling of neuronal arbors.
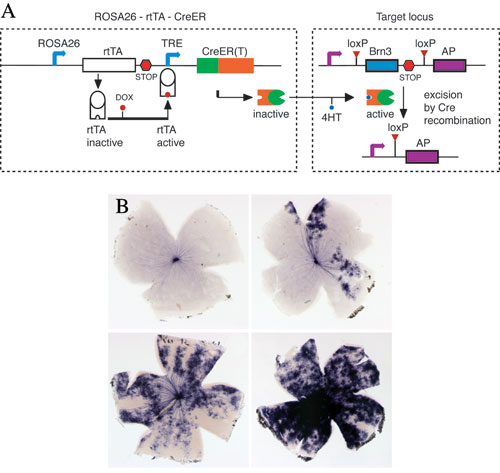
A. Dual pharmacological control of Cre-loxP recombination: In the ROSA26-rtTA-CreER knock-in line, rtTA is constitutively transcribed under the control of the ROSA26 promoter, activated by the addition of Doxycycline, binds the tetracycline response element (TRE) and induces transcription of CreER. CreER is then activated by 4HT, to induce recombination at target loci, with excision of an endogenous gene and expression of a reporter.
B. Retinal flat mounts from mice exposed to increasing amounts of Doxycycline and 4HT show increased frequency of recombination.
Defining the transcriptional requirements for the formation of Retinal Ganglion Cell (RGC) types.
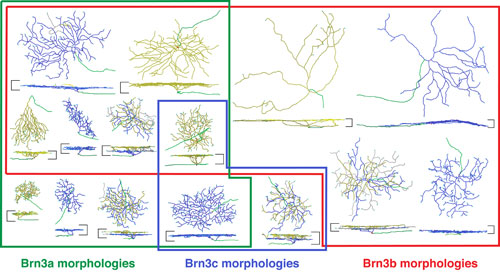
Morphologies of mouse retinal ganglion cells visualized by sparse genetic activation of an alkaline phosphatase reporter targeted to the Brn3a, Brn3b, or Brn3c loci. Retinal ganglion cells were traced using Neuromantic software and are shown en face and in cross section. Colored boundaries indicate the morphologic types characteristic for retinal ganglion cells expressing the indicated Brn3 family member.
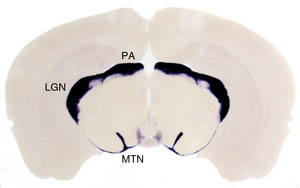
Coronal section through the brain of a Brn3aCKOAP/+; Pax6 :Cre mouse. Retinorecipient areas receiving projections from RGCs expressing Brn3a are labeled in purple, and include the Medial Terminal Nucleus (MTN) of the Accessory Optic System, the Lateral Geniculate Complex (LGN) and the Pretectal Area (PA).
Dissecting the molecular mechanisms of dendritic arbor formation and axon guidance in RGCs.
Determining the specific functions of RGCs in visual circuits under normal and pathologic conditions. By specifically ablating individual cell types and investigating visual behaviors/reflexes in live animals or RGC responses to visual stimuli in mutant retinas, we hope to uncover both the normal functions of RGCs in specific visual circuits, as well as define the neurological defects resulting from pathological changes in various elements of the circuit.
Selected Publications
Badea TC, Williams J, Smallwood P, Shi M, Motajo O, Nathans J. Combinatorial expression of Brn3 transcription factors in somatosensory neurons: genetic and morphologic analysis. J Neurosci. 2012 Jan 18;32(3):995-1007. PubMed
Matsuoka RL, Chivatakarn O, Badea TC, Samuels IS, Cahill H, Katayama K, Kumar SR, Suto F, Chédotal A, Peachey NS, Nathans J, Yoshida Y, Giger RJ, Kolodkin AL. Class 5 transmembrane semaphorins control selective Mammalian retinal lamination and function. Neuron. 2011 Aug 11;71(3):460-73.
Chen SK, Badea TC (co-corresponding author), Hattar S. Photoentrainment and pupillary light reflex are mediated by distinct populations of ipRGCs. Nature. 2011 Jul 17;476(7358):92-5.
McNeill DS, Sheely CJ, Ecker JL, Badea TC, Morhardt D, Guido W, Hattar S. Development of melanopsin-based irradiance detecting circuitry. Neural Dev. 2011Mar 18;6(1):8. [Epub ahead of print]
Matsuoka RL, Nguyen-Ba-Charvet KT, Parray A, Badea TC, Chedotal A, Kolodkin AL. Transmembrane semaphorin signalling controls laminar stratification in the mammalian retina. Nature. 2011 Feb 10;470(7333):259-63.
Badea TC, Nathans J. Morphologies of mouse retinal ganglion cells expressing transcription factors Brn3a, Brn3b, and Brn3c: analysis of wild type and mutant cells using genetically-directed sparse labeling. Vision Res. 2011 Jan 28;51(2):269-79. Epub 2010 Sep 6.
Badea TC, Hua ZL, Smallwood PM, Williams J, Rotolo T, Ye X, Nathans J. New mouse lines for the analysis of neuronal morphology using CreER(T)/loxP-directed sparse labeling. PLoS One. 2009 Nov 16;4(11):e7859.
Ye X, Wang Y, Cahill H, Yu M, Badea TC, Smallwood PM, Peachey NS, Nathans J. Norrin, frizzled-4, and Lrp5 signaling in endothelial cells controls a genetic program for retinal vascularization. Cell. 2009 Oct 16;139(2):285-98. Erratum in: Cell. 2010 Apr 2;141(1):191.
Badea TC, Cahill H, Ecker J, Hattar S, Nathans J. Distinct roles of transcription factors brn3a and brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron. 2009 Mar 26;61(6):852-64.
Guler AD, Ecker JL, Lall GS, Haq S, Altimus CM, Liao HW, Barnard AR, Cahill H, Badea TC, Zhao H, Hankins MW, Berson DM, Lucas RJ, Yau KW, Hattar S. Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature. 2008 May 1;453(7191):102-5. Epub 2008 Apr 23.
Badea TC, Nathans J. Quantitative analysis of neuronal morphologies in the mouse retina visualized by using a genetically directed reporter. J Comp Neurol. 2004 Dec 20;480(4):331-51.
Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci. 2003 Mar 15;23(6):2314-22.
Structure

| Name | Title | |
|---|---|---|
Tudor Constantin Badea, M.D, Ph.D. |
Unit Head | badeatc@mail.nih.gov |
| Friedrich Kretschmer | Postdoctoral Fellow | friedrich.kretschmer@nih.gov |
| Miruna Ghinia | Predoctoral Fellow | miruna.ghinia@nih.gov |
| Szilard Sajgo | Predoctoral Fellow | szilard.sajgo@nih.gov |
| Nadia Parmhans | Intern | nadia.parmhans@nih.gov |
Alumni /Former Lab Members
- Sumit Kumar
- Oluwaseyi Motajo
- Melody Shi
Last Reviewed: May 2012

